De Novo Peptide Sequencing Services
Peptides are compounds formed by α-amino acids linked together via peptide bonds. They are widely found in animals, plants, and microorganisms, playing biochemical roles as signaling molecules, toxins, cytokines, and more. Peptides are also fundamental building blocks of proteins, playing critical roles in various biological systems. Digesting proteins into peptides has become a standard method for protein identification and quantification. Peptide sequencing technology has undergone rapid advancements over the years. Edman degradation, the earliest peptide sequencing method, has limitations such as low throughput, low sensitivity, and inability to directly analyze peptide mixtures or peptides with blocked termini. Conventional proteomics methods are unsuitable for identifying peptides that do not exist in known databases. For unknown species, novel post-translational modifications, or amino acid mutations, database-dependent methods are often ineffective in identifying peptide sequences. De novo peptide sequencing services provide a revolutionary solution by directly determining the amino acid sequence of peptides without relying on any known sequence or protein database information. This services hold great potential for identifying entirely novel peptides and peptides with unclear protein origins.
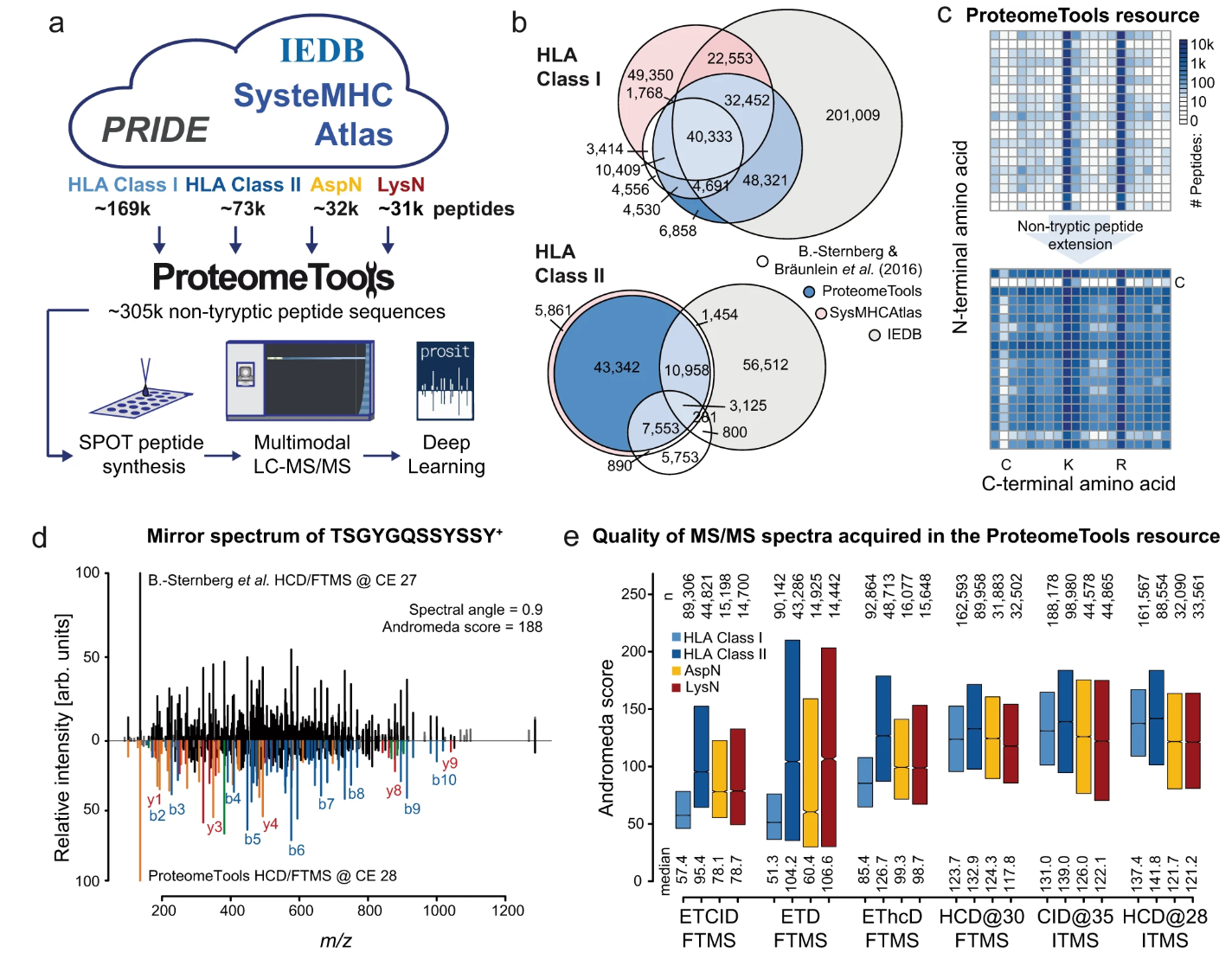
Wilhelm, M. et al. Nature Communications, 2021.
Figure 1. One Application of De Novo Peptide Sequencing: Immunopeptidome/Tumor Neoantigen Discovery.
Services at MtoZ Biolabs
De novo peptide sequencing using mass spectrometry involves collecting the mass information of amino acid sequences in peptide fragments (MS/MS spectra) and deciphering peptide sequences directly from the spectra using de novo sequencing algorithms. Utilizing Thermo Fisher's Orbitrap series mass spectrometers in combination with a Nano-LC system, MtoZ Biolabs has established an advanced platform for de novo peptide sequencing. Our comprehensive de novo peptide sequencing services encompass all steps, including sample preparation → peptide separation → mass spectrometry detection → data analysis.
MtoZ Biolabs has developed proprietary software for de novo sequencing analysis, leveraging its extensive expertise in mass spectrometry. Our software features custom algorithms for processing raw mass spectrometry data, adhering to a broad-input, narrow-output principle. This ensures comprehensive data inclusion while enabling precise peptide sequence identification. Our de novo peptide sequencing services employ algorithms based on advanced AI-driven large language models to directly decode amino acid composition and sequence from MS/MS spectra. These cutting-edge algorithms allow us to distinguish amino acids with high accuracy and perform quantitative analysis of peptide abundance within protein samples. The de novo peptide sequencing services provided by MtoZ Biolabs can accurately analyze the amino acid composition, sequence and abundance of peptides, and provide accurate peptide results.
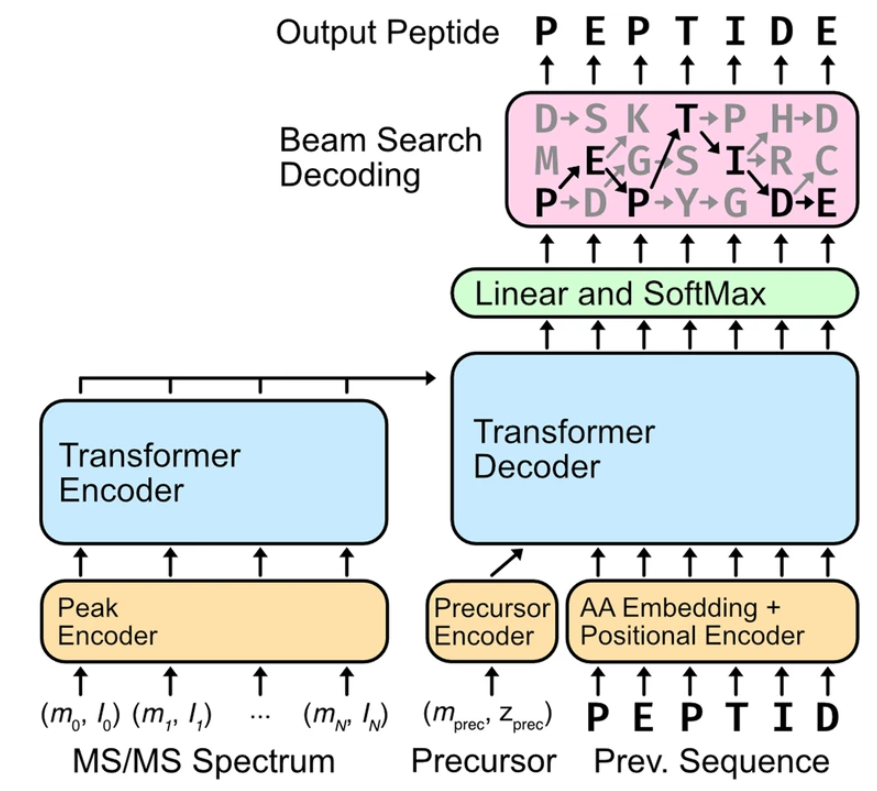
Yilmaz, M. et al. Nature Communications, 2025.
Figure 2. Overview of De Novo Sequencing Methods Based on Artificial Intelligence Large Language Model.
Service Advantages
1. Advanced Analytical Platform
MtoZ Biolabs has established advanced de novo peptide sequencing services platform, ensuring reliable, rapid, and high-precision analytical services.
2. Transparent Pricing
Our pricing structure is transparent, with no hidden fees or additional fees.
3. High Data Quality
Data depth coverage and strict data quality control. AI-driven platforms ensure the accuracy of sequencing and provide customers with comprehensive data reports.
Sample Submission Suggestions
1. Sample Type
Peptide sample quantity must exceed 5 micrograms.
Note: If you have special requirements or your samples do not meet the above criteria, please contact us.
Applications
1. Immunopeptidomics
De novo peptide sequencing services can identify immune peptides presented by MHC molecules.
2. Tumor Neoantigen Discovery
De novo peptide sequencing technology can identify tumor-specific mutated peptides.
3. Metaproteomics
In cases where microbial sequence databases are incomplete, de novo peptide sequencing services can uncover novel peptides within microbial communities.
4. Peptide Mutation Detection
Amino acid mutations in peptides can significantly impact product stability and efficacy. De novo sequencing enables identification of mutation sites and assessment of mutation levels.
5. Discovery of Functional Peptides
De novo peptide sequencing technology can assist in the discovery of new antimicrobial peptides, active peptides and so on.
Case Study
1. Discovery of Immune Peptides Using De Novo Sequencing
A de novo mass spectrometry sequencing approach, coupled with an MHC-centered analysis strategy, was employed to detect cancer-specific non-classical MHC immune peptides without prior acquisition of target sequences from genomic or RNA sequencing data. This study reanalyzed high-quality datasets of non-classical MHC immune peptides from human cancers within a large synthetic peptide library to detect and validate of lncRNA-derived peptides in human cervical tumor resection. Mass Spectrometry de novo peptide sequencing services are designed to discover novel, immunogenic, non-classical peptide sequences in primary tumor tissue.
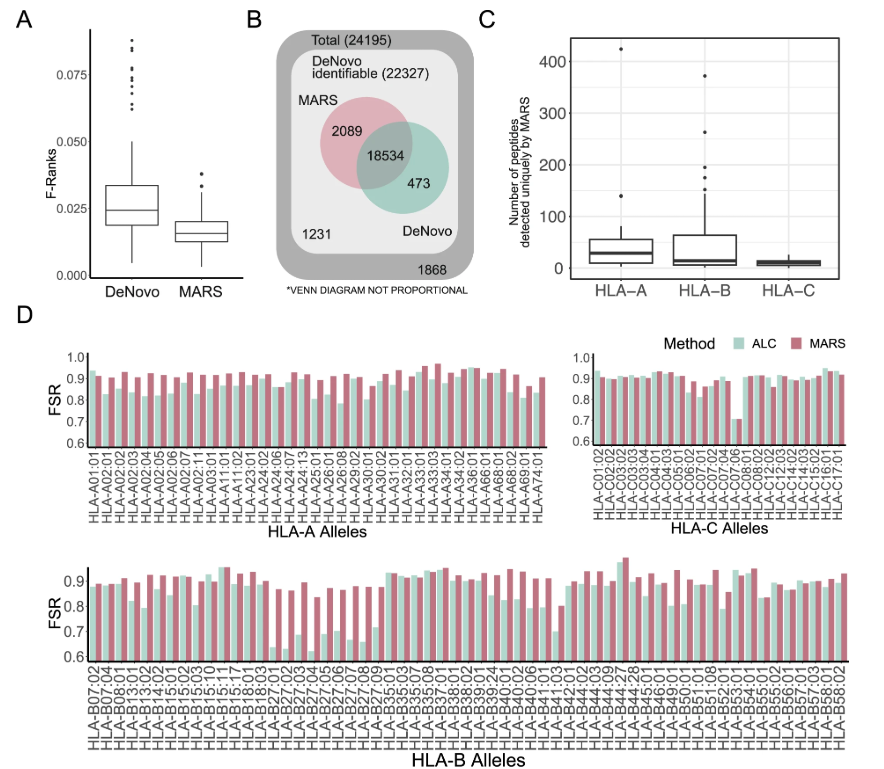
Liao, HQ. et al. Nature Communications, 2024.
Figure 3. De Novo Sequencing for the Discovery of Novel Immune Peptides.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Quality Control Assessment
4. Peptide Sequence Tables
5. Statistical Information of Peptide Sequences
6. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







