2D DIGE Analysis Service
2D DIGE Analysis Service uses fluorescent labeling technology to separate and compare proteins from multiple samples on the same gel, which can avoid quantitative errors caused by gel-to-gel variation and improving the sensitivity and accuracy of quantitative proteomic analysis. Two-Dimensional Difference Gel Electrophoresis (2D DIGE) is a high-resolution protein separation and quantitative analysis method based on traditional two-dimensional electrophoresis. It separates protein mixtures labeled with fluorescent dyes (e.g., Cy2, Cy3, Cy5) according to differences in isoelectric points and molecular weights. Fluorescence signals from the samples are captured by a fluorescence scanner, and specialized software is used to quantitatively compare protein expression levels. Due to its high sensitivity, strong resolution, and excellent quantitative accuracy, 2D DIGE is widely applied in the study of differential protein expression. MtoZ Biolabs provides 2D DIGE Analysis Service and combines chromatography and mass spectrometry platforms to provide customers with high-quality proteomics analysis service.
Analysis Workflow
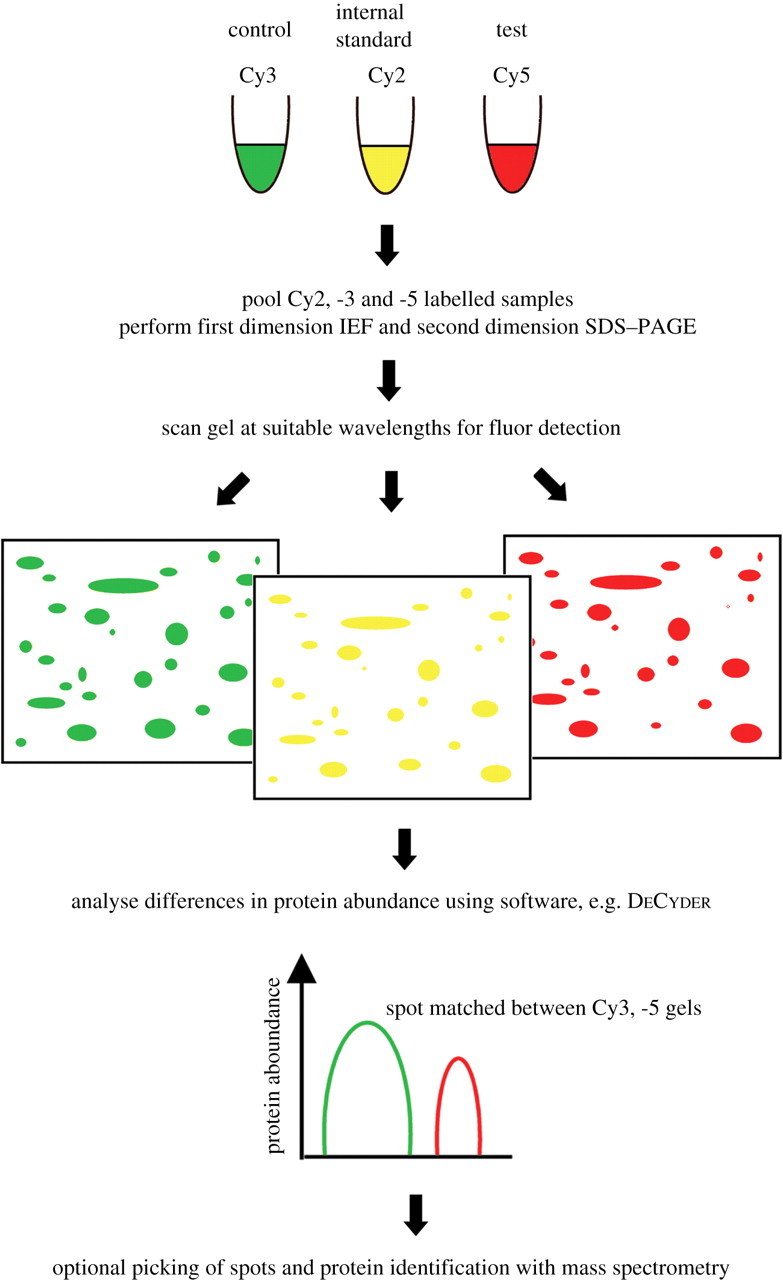
Mcnamara L E. Journal of the Royal Society Interface. 2010.
1. Sample Preparation
Extract proteins and remove interfering substances.
Accurately quantify protein concentration to ensure consistent labeling efficiency.
2. Fluorescent Dye Labeling
Label control and experimental group samples with CyDye fluorescent dyes, then mix and load onto the same gel.
3. Two-Dimensional Electrophoresis
Separate proteins in the sample using isoelectric focusing (IEF) followed by SDS-PAGE.
4. Image Acquisition and Data Analysis
Capture images using a fluorescence scanner and perform quantitative analysis with specialized software to identify differentially expressed protein spots.
5. Differential Protein Identification (Optional)
Excise significantly different protein spots for further identification and functional analysis using mass spectrometry.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs offers 2D DIGE Analysis Service along with comprehensive quantitative proteomics analysis integrated with advanced chromatography and mass spectrometry technologies.
Applications
Differential Protein Screening
2D DIGE analysis service can be used to compare protein expression differences between disease and normal states to identify potential biomarkers.
Drug Mechanism Research
Evaluate the impact of drugs on protein expression profiles in cells or tissues to uncover mechanisms of action.
Signal Pathway Research
2D DIGE analysis service can be used to analyze expression changes of pathway-related proteins under specific conditions.
Environmental and Stress Research
Investigate proteomic responses to environmental changes or stress conditions, revealing biological adaptation mechanisms.
FAQ
Q. How to Prevent Salt or Detergent Interference During Fluorescent Labeling in 2D DIGE Analysis?
1. Select Appropriate Protein Extraction Buffer
Use low-salt buffers to extract proteins, avoiding high-salt environments that inhibit the CyDye labeling reaction. Remove unnecessary detergents (e.g., SDS) or high concentrations of urea to ensure buffer compatibility with labeling conditions.
2. Dialysis or Ultrafiltration to Remove Interfering Substances
Perform repeated dialysis of the protein solution with a low-salt buffer (e.g., Tris-HCl) to remove small molecule salts. Alternatively, concentrate the protein solution through an ultrafiltration tube to remove salt and small molecule impurities at the same time.
3. Clear Detergents from Protein Samples
Precipitate proteins using acetone or methanol-chloroform to remove detergents such as SDS.
Use specialized cleanup kits for further sample purification.
4. Optimize Labeling Reaction Conditions
Ensure the final salt concentration of the protein sample is below 10 mM, and the pH is between 8.5 and 9.0 to maximize CyDye labeling efficiency.
Q. How to Select a Suitable Internal Standard (Cy2-Labeled Sample) to Correct Experimental Errors in 2D DIGE Analysis?
1. Composition of the Internal Standard
Mixed Sample: Combine equal amounts of protein from all experimental and control group samples to create an internal standard. A mixed sample represents the overall protein expression profile, suitable for correcting gel-to-gel experimental variations.
Representative Sample: If a mixed sample is not feasible, choose a single representative sample with a protein expression profile close to the overall sample profile as the internal standard.
2. Labeling Quantity of the Internal Standard
According to the labeling ratio of CyDye, ensure that the concentration of the internal standard labeled with Cy2 is sufficient and consistent with the signal intensity of the Cy3 and Cy5 labeled samples.
3. Reproducibility Validation of the Internal Standard
Load the same Cy2-labeled internal standard on multiple gels, and verify the consistency of its fluorescence intensity and spot positions to ensure stability and comparability.
Case Study
This study employed 2D DIGE analysis to reveal that low-temperature stress significantly altered the expression levels of various metabolism-related proteins, particularly key enzymes involved in glycolysis, lipid metabolism, and the TCA cycle. Furthermore, mass spectrometry identification uncovered specific post-translational modification sites, such as phosphorylation, in low-temperature-responsive proteins, shedding light on the critical roles these modifications play in regulating enzymatic activity and metabolic adaptation.
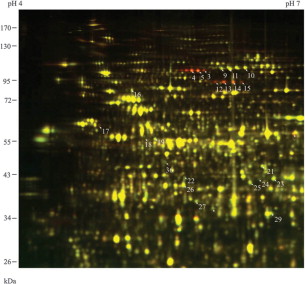
Li T. et al. Molecular Plant. 2011.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
2D-DIGE Based Protein Quantitative Service
How to order?







