Single Cell Whole Genome Sequencing Service
Single cell whole genome sequencing (scWGS) is a technology used to analyze the genetic information of individual cells by sequencing their entire genome. Single cell whole genome sequencing service overcomes the challenge of effectively handling complex cellular heterogeneity, a limitation of traditional whole genome sequencing techniques, by amplifying and performing high-throughput sequencing on the entire genome of each individual cell. Single cell whole genome sequencing has been widely applied in various fields, due to its accuracy and high resolution. For example, in tumor research.Tumor cells typically exhibit heterogeneity, and single cell whole genome sequencing can assist researchers in revealing genomic mutations, copy number variations, and gene rearrangements within tumors, particularly aiding in the understanding of tumor evolution and mechanisms of drug resistance.
Single cell whole genome sequencing service generally combine microfluidic technology, single cell isolation techniques, amplification methods (such as MDA or DOP-PCR), and high-throughput sequencing platforms (such as Illumina), thereby enabling the acquisition of genome-wide data from individual cells while maintaining high coverage. MtoZ Biolabs is dedicated to providing advanced and precise single cell whole genome sequencing services assisting you in gaining deep insights into how genomic variations, chromosome rearrangements, and other subtle genomic structural changes manifest within individual cells.
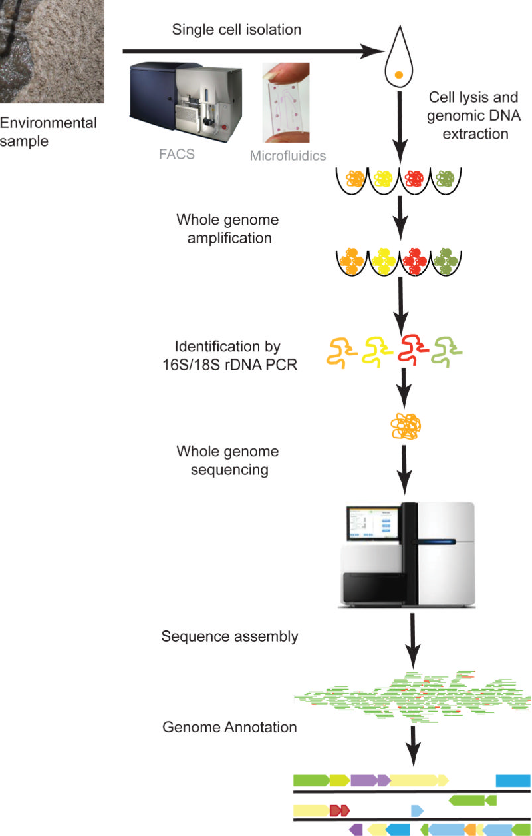
Yilmaz, S. et al. Current opinion in biotechnology. 2012.
Analysis Workflow
The workflow of single cell whole genome sequencing typically involves several steps, including sample preparation, single cell isolation, genome amplification, and high-throughput sequencing.
Sample Preparation
First, individual cells are isolated from tissues or cultures, ensuring the integrity and representativeness of the sample. MtoZ Biolabs utilizes advanced microfluidic sorting and other technologies to minimize cell damage.
Single cell isolation and amplification
Each single cell is individually allocated to a microreactor for DNA extraction and amplification. Efficient techniques such as Multiple Displacement Amplification (MDA) or DOP-PCR are utilized to ensure comprehensive amplification of the genomic information extracted from the single cell.
High-throughput sequencing
The amplified DNA samples undergo high-throughput sequencing using platforms like Illumina for deep sequencing, ensuring comprehensive coverage and accuracy of each cell's genome.
Data Analysis and Interpretation
The sequencing data is processed using advanced bioinformatics analysis platforms, including data quality control, genome reconstruction, and variant detection. MtoZ Biolabs provides customized analysis reports to help clients gain a deep understanding of the key data in their research.
Case Study
Single cell whole genome sequencing revealed somatic mutations accumulate with age in human B lymphocytes.
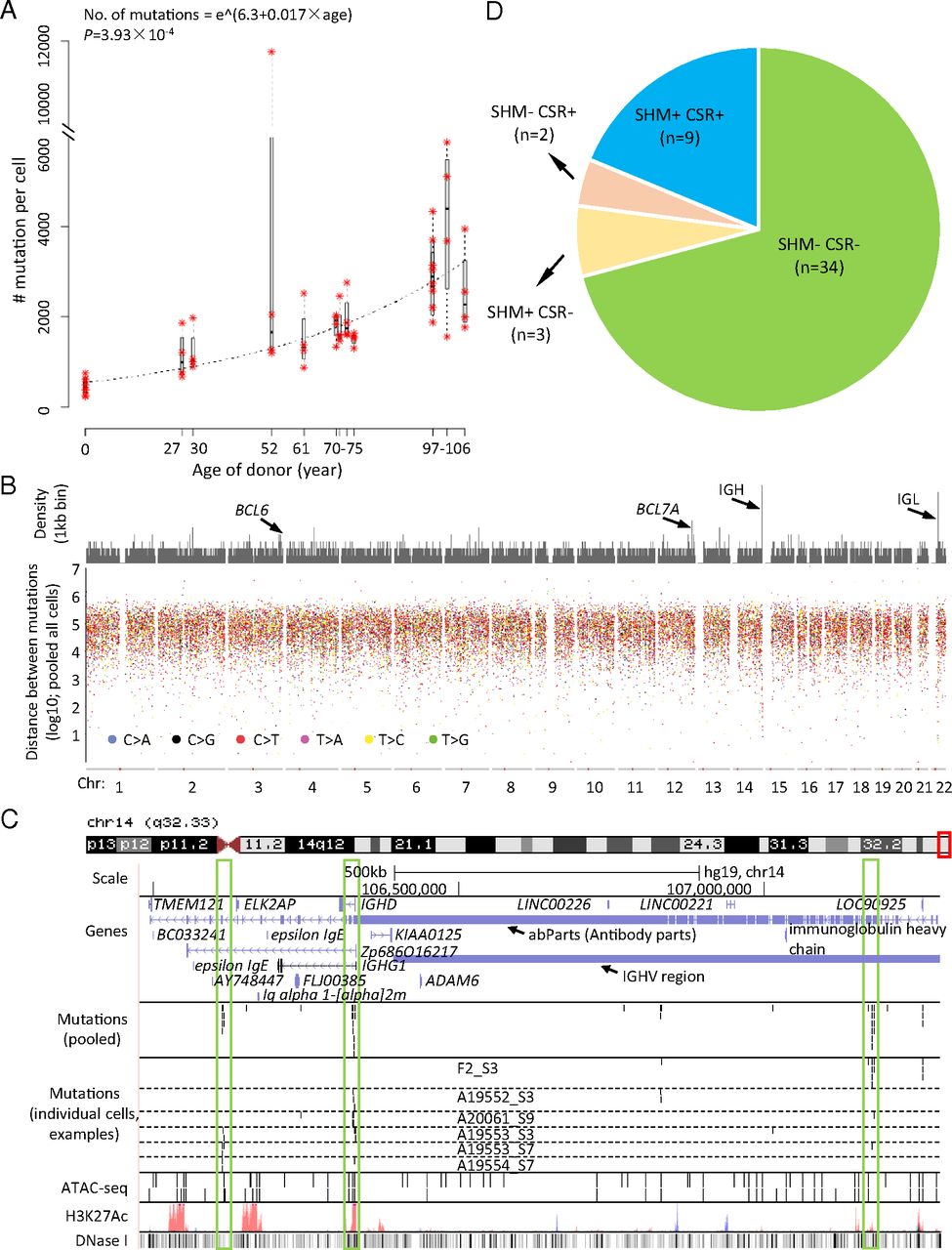
L. Zhang. et al. PNAS. 2019.
Sample Submission Suggestions
Sample Types
We accept single cell samples from various sources, including but not limited to tumor tissues, peripheral blood, embryonic cells, and individual cell lines. Each sample must contain a sufficient number of cells for single cell isolation.
Sample quantity
The quality of the samples and the viability of the cells directly impact the quality of the data. Therefore, it is recommended to contact us to consult on sample quantity before submission.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
FAQ
Q: Efficiency and Purity of Cell Isolation: How to ensure that isolated single cells maintain sufficient purity and avoid contamination from cell populations? How can the efficiency of single cell isolation be improved for low-abundance cell types?
The efficiency and purity of the single cell isolation process directly affect the subsequent sequencing quality and data accuracy. Below are several strategies to ensure the purity of single cell isolation:
Flow cytometry (FACS): Flow cytometry utilize fluorescent markers for precise cell sorting. By employing cell surface markers (such as antibody labeling), target cells can be efficiently sorted, ensuring high purity of the single cells. For low-abundance cell types, the high sensitivity and customizability of flow cytometry make it an ideal choice.
Microfluidic technology: This technology captures and immobilizes individual cells using a microchannel system, which can significantly enhance separation efficiency, especially in high-throughput single cell analysis.Microfluidic technology separates cells based on size, morphology, and other physical characteristics.Therefore, its advantages in high throughput and high resolution are particularly prominent when handling low-abundance cells.
Contamination-Controlled Experimental Environment: During the single cell isolation process, cross-contamination during handling may lead to contamination of the cell population. To minimize such contamination, it is essential to rigorously control the experimental environment, use sterile platforms and tools, and implement strict quality control measures on the samples to ensure that the entire process, from isolation to capture, is free of contamination.
Dual/Triple Separation Strategy: For low-abundance cells, the separation efficiency and purity can be ensured by combining multiple techniques, such as FACS, microfluidics, and single cell clonal amplification.The use of multiple markers can improve the ability to identify low-abundance cells, thereby increasing the success rate of cell isolation.
How to order?







