Renal Disease Diagnosis-Applied Exosomes
Renal Disease Diagnosis-Applied Exosomes aims to analyze exosomes derived from patients with renal diseases using exosome-based analytical techniques, in order to identify disease-related biomarkers, supporting early diagnosis, pathological assessment, disease monitoring, and treatment efficacy evaluation for renal conditions.
Renal diseases are a major global health concern, particularly chronic kidney disease (CKD) and acute kidney injury (AKI). Early diagnosis and accurate monitoring are critical for effective treatment and improved prognosis. Traditional diagnostic methods, such as serum creatinine and urinary protein detection, are widely used in clinical practice but often lack sensitivity in early stages and may fail to provide a comprehensive assessment of the extent and nature of kidney injury in certain cases.
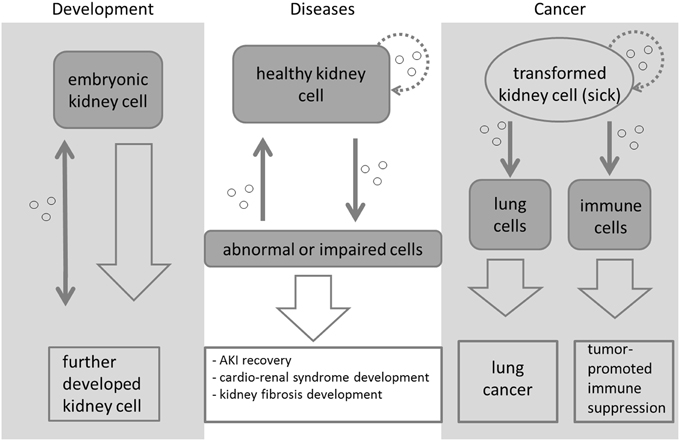
Mirja K. et al. Frontiers in Cell and Developmental Biology. 2015.
Figure 1. Exosome-mediated signaling during kidney development, disease, and cancer.
As nanoscale vesicles secreted by cells, exosomes carry a rich array of biomolecules, including proteins, lipids, and RNAs, and play critical roles in disease progression. In recent years, exosomes and their components have drawn significant attention as non-invasive biomarkers for kidney diseases. Studies have shown that proteins and miRNAs contained in urinary exosomes are involved in intercellular communication within the urinary microenvironment and are closely associated with the onset and progression of renal diseases.
Leveraging advanced multi-omics analysis platforms, MtoZ Biolabs' Renal Disease Diagnosis-Applied Exosomes service offers comprehensive exosome isolation and analysis solutions, to help clients uncover valuable biomarker information from exosomes, supporting early diagnosis, therapeutic evaluation, and personalized treatment of kidney diseases.
Analysis Workflow
1. Exosome Isolation and Quality Control
High-purity exosomes are isolated using multiple methods, followed by rigorous quality assessment to ensure sample integrity.
2. Extraction of Target Biomolecules
Based on project requirements, target biomolecules are extracted from the exosomes.
3. Biomarker Detection
Proteomics, metabolomics, or high-throughput sequencing techniques are used to analyze the extracted biomolecules.
4. Data Analysis and Reporting
Comprehensive data interpretation and bioinformatics analysis are performed, including identification and functional analysis of disease-related biomarkers, with a detailed experimental report provided.
Applications
Examples of applications for Renal Disease Diagnosis-Applied Exosomes:
Chronic Kidney Disease (CKD) Monitoring
miRNAs, mRNAs, and proteins within exosomes reflect kidney injury and fibrosis. Exosome analysis enables detection of early markers of kidney damage, providing a non-invasive tool for monitoring CKD progression.
Acute Kidney Injury (AKI) Diagnosis
AKI presents as sudden renal dysfunction, while traditional creatinine testing often lags behind pathological changes. Exosome analysis offers a non-invasive diagnostic method for early-stage AKI.
Kidney Fibrosis Detection
Kidney fibrosis is a hallmark of CKD progression. Exosome profiling provides dynamic insights into fibrotic processes and helps evaluate therapeutic efficacy.
Renal Transplant Rejection Monitoring
Immune rejection following kidney transplantation is a critical factor affecting transplant success and long-term survival. Exosome analysis enables biomarker discovery and screening for non-invasive monitoring of rejection episodes.
FAQ
Q. What is the relationship between exosomes and renal disease diagnosis?
Exosomal biomolecules reflect the physiological state of their parent cells. In the diagnosis of renal diseases, analyzing the biomolecular content of exosomes in body fluids such as blood, urine, or renal biopsy fluid can provide valuable insights into kidney function and pathology.
Q. How is the purity and quality of exosomes ensured to make them suitable for renal disease research?
The purity and quality of exosomes are critical for renal disease studies. The following methods are used to ensure high-quality exosomes:
1. Exosome Isolation Methods: Techniques such as ultracentrifugation, size-exclusion chromatography (SEC), and immunoaffinity capture are employed.
2. Quality Control: Transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) are used to confirm exosome morphology and concentration. Western blot is performed to detect exosomal markers (e.g., CD9, CD63) and ensure the removal of contaminants.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study primarily investigated the roles of mitochondrial tRNAs and miRNAs in exosomes during inflammation of renal tubular epithelial cells. Researchers employed miRNA sequencing to compare the expression differences of exosomal miRNAs between healthy controls and renal disease models. The results revealed that under inflammatory stimulation, exosomes secreted by renal tubular epithelial cells exhibited significant expression changes in 30 miRNAs and 16 mitochondrial RNAs, suggesting that mitochondria play a critical role in sensing renal inflammation. These findings indicate that these exosomal non-coding RNAs (ncRNAs) may serve as potential indicators for predicting the pathogenesis of chronic kidney disease and for monitoring therapeutic response.
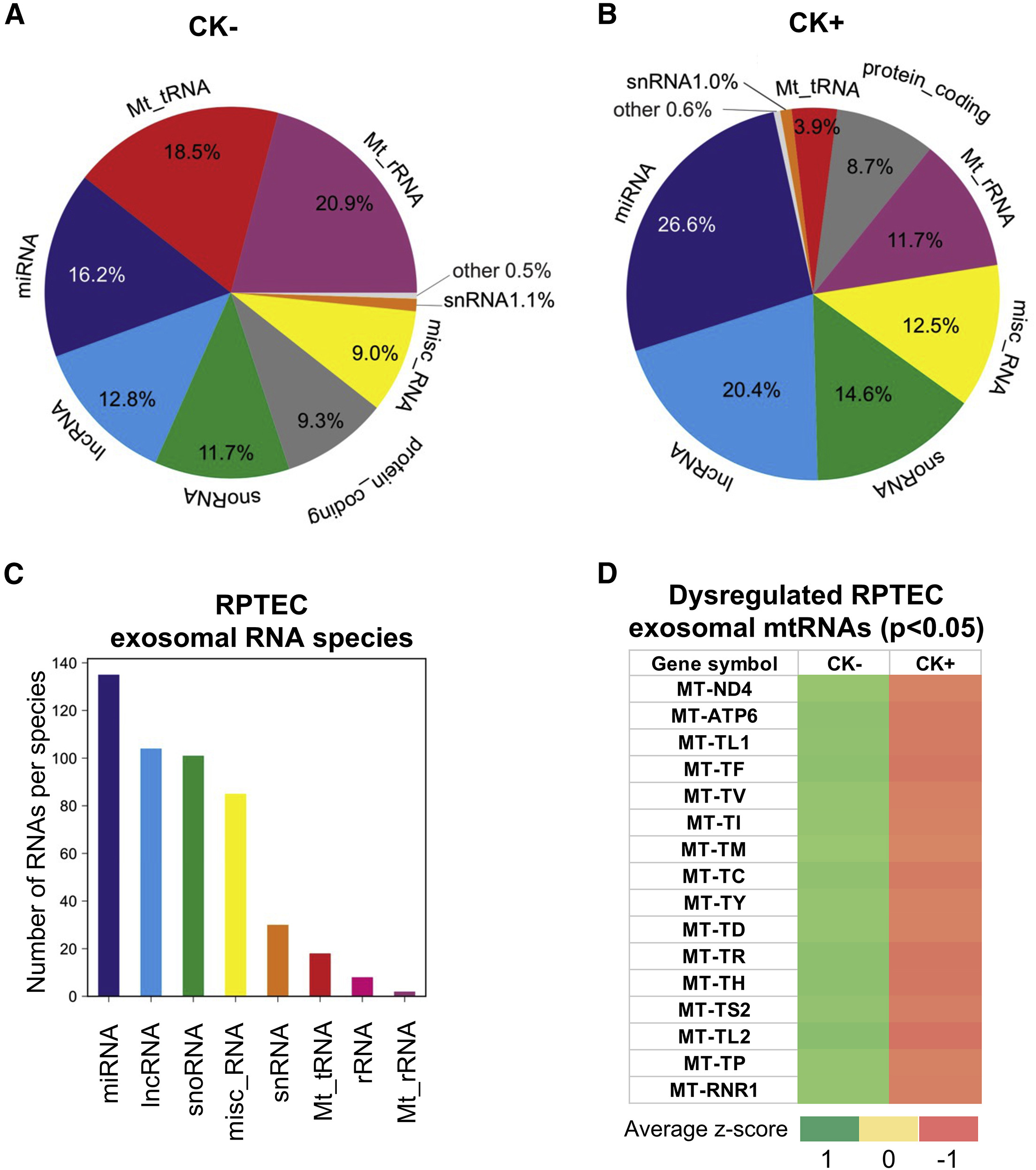
Ranches G. et al. Molecular Therapy Nucleic Acids. 2022.
Figure 2. Identification of dysregulated exosomal ncRNAs from a cell-based model system of renal inflammation by RNA sequencing.
How to order?







