Metabolic Pathway Analysis Service
- Hypergeometric Distribution: It is suitable for small-scale metabolite set analysis, identifying significant pathways through enrichment analysis, and the calculation is intuitive and fast.
- GSEA (Gene Set Enrichment Analysis): Ideal for large-scale metabolomics data, focusing on overall pathway activity changes, especially for dynamic or continuous data.
- PLS-DA (Partial Least Squares Discriminant Analysis): Best suited for distinguishing metabolic features between experimental groups. It can be combined with pathway enrichment to interpret inter-group differences.
Metabolic pathway analysis service integrates metabolomics and bioinformatics approaches to systematically analyze the dynamic changes of metabolites within biological systems and their regulatory pathways. This service not only identifies key molecules in metabolic networks but also links metabolic activities to biological functions, disease mechanisms, and drug actions, providing researchers with deeper insights into complex biological systems. Leveraging advanced metabolomics platforms and precise pathway analysis tools, MtoZ Biolabs offers comprehensive metabolic pathway analysis service from metabolite detection to pathway interpretation, providing essential support for basic research, disease mechanism elucidation, and drug development.
Chen Y. et al. Metabolites. 2022.
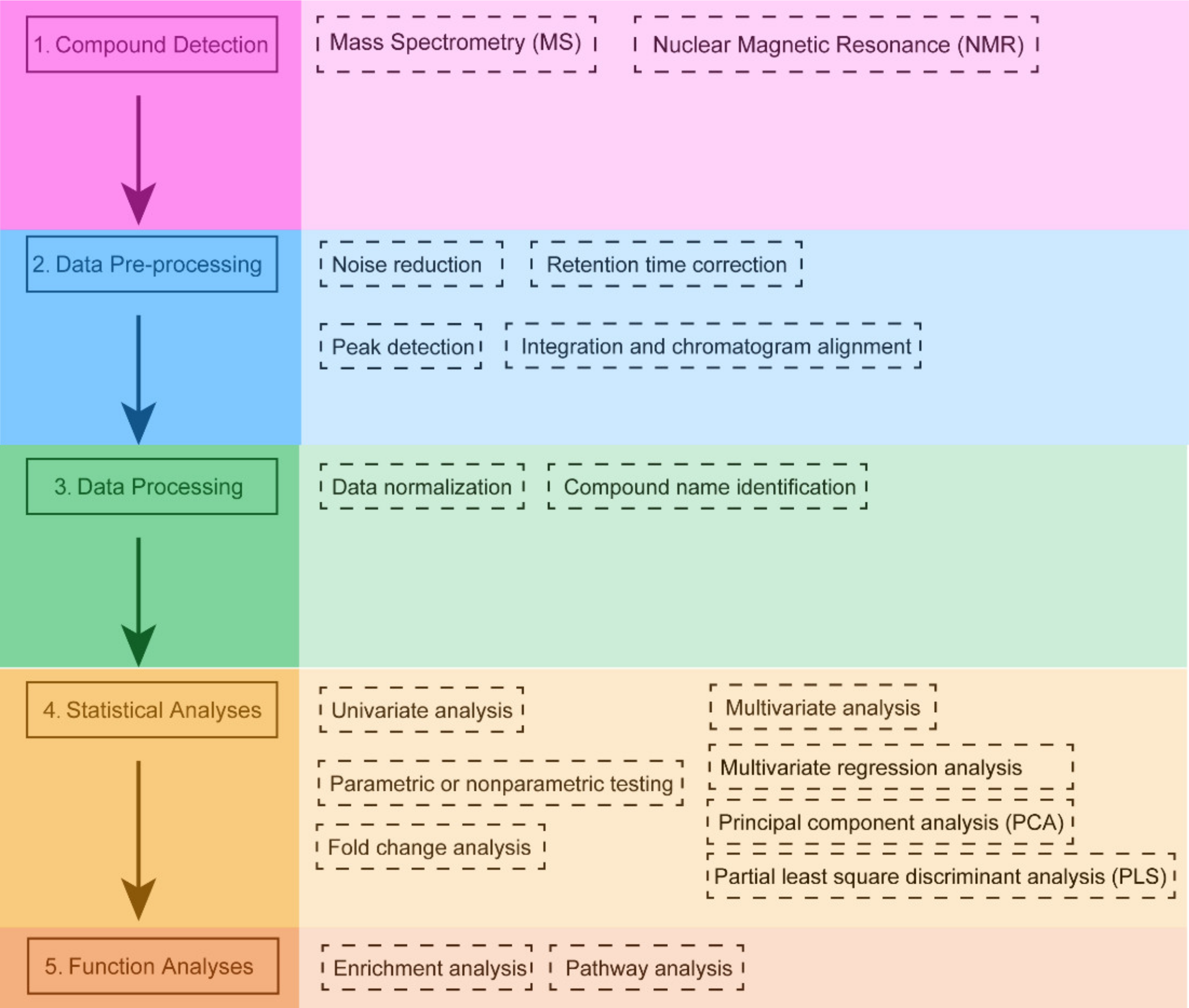
Analysis Workflow
1. Sample Preparation and Metabolite Detection
Metabolites are extracted based on the sample type. Techniques such as LC-MS, GC-MS, or NMR are used to analyze metabolites and generate metabolomics data.
2. Data Preprocessing
Collected data undergo preprocessing steps, including noise reduction, peak identification, standardization, and normalization.
3. Metabolite Identification and Quantification
Metabolites are identified using established databases such as HMDB and KEGG. Relative abundances of metabolites are quantified using standard curves or mass spectrometry data.
4. Pathway Mapping and Enrichment Analysis
Metabolite data is matched with pathway databases (e.g., KEGG, Reactome, MetaCyc) to identify and associate key metabolic reactions and pathways. Statistical methods and computational tools are employed to perform pathway enrichment analysis and highlight significantly altered metabolic pathways.
5. Result Interpretation and Report Generation
Comprehensive pathway maps and enrichment analysis results are produced, accompanied by biological interpretation to provide meaningful insights.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced metabolic pathway analysis platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all metabolic pathway analysis data, providing clients with a comprehensive data report.
4. Customized Analysis Plans: Tailored metabolic pathway analysis strategies are designed based on client requirements to meet diverse research needs.
Applications
Disease Research
Uncover metabolic pathway abnormalities in complex diseases such as cancer, diabetes, and cardiovascular diseases.
Drug Development
Analyze the effects of drugs on metabolic pathways, identify potential therapeutic targets, and evaluate side effects.
Precision Medicine
Provide metabolomics data to support personalized diagnosis and treatment.
Nutrition and Metabolism Research
Examine the impact of diet on metabolic activities and investigate regulatory mechanisms of metabolic health, supporting precision nutrition studies.
Environmental Metabolomics Research
Study the effects of environmental factors (e.g., toxins, pollutants) on biological metabolism and their molecular mechanisms.
FAQ
Q. How to choose statistical methods (e.g., hypergeometric distribution, GSEA, or PLS-DA) to identify significantly altered metabolic pathways?
The choice depends on the data scale, experimental design, and analytical objectives.
Q. How can metabolic pathway alterations be linked to specific biological questions (e.g., disease mechanisms or drug effects)?
Significant pathway changes can be mapped to established biological networks (e.g., KEGG or Reactome) to identify core pathways related to the target disease or drug response. Additionally, the functional annotations of upstream and downstream metabolites and published literature are combined to infer the connection between the pathway and specific physiological or pathological processes. Finally, experimental validation of key metabolites reinforces the link between pathways and the biological questions under investigation.
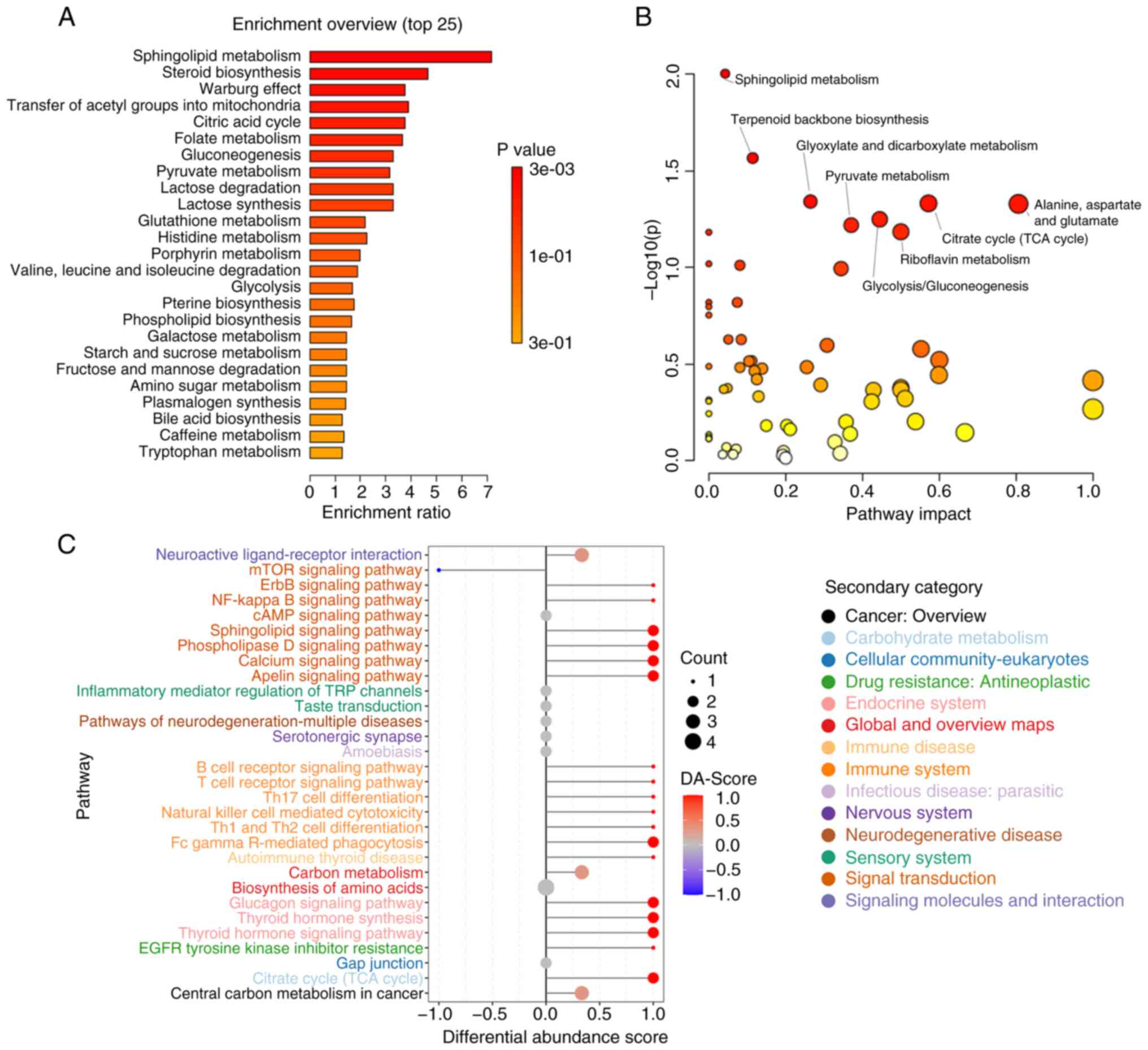
Case Study
This study employed serum metabolomics analysis to investigate the metabolic characteristics of patients with chronic obstructive pulmonary disease (COPD), particularly those with frequent exacerbation phenotypes. Metabolic pathway analysis revealed significant disruptions in metabolic pathways, including energy metabolism (e.g., tricarboxylic acid cycle), amino acid metabolism (e.g., branched-chain amino acid metabolism), and lipid metabolism. These alterations were closely associated with inflammation, oxidative stress, and immune regulation. The findings suggest that metabolic pathway dysregulation may underlie the frequent exacerbation phenotype, providing novel insights and molecular evidence for personalized treatment strategies and biomarker development in COPD.
Ding H Z. et al. Molecular Medicine Reports. 2024.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
How to order?







