Drug Affinity Responsive Target Stability Analysis Service
Drug affinity responsive target stability (DARTS) is a label-free small molecule (SM) probe technique under chemical proteomics first proposed by Lomenick et al. in 2009, which makes it possible to track and identify target proteins whose stability changes after binding with small molecules (SMs). To date, DARTS has been a focus in tracking proteins in animals, fungi, bacteria, and plants, and is widely applied with high simplicity, efficiency, universality, and accuracy.
The principle of DARTS is: under physiological conditions, proteins are in dynamic equilibrium with a variety of available conformations and may exhibit different degree of local reversible unfolding. When a specific ligand reaches saturation, due to the negative free energy change caused by hydrophobic, hydrogen bonding, and/or electrostatic interactions formed between the protein and the drug ligand, the equilibrium will shift, thus highly favoring the ligand-binding conformation. This forms a thermodynamically more stable state, in which the fluctuations and unfolding of the target protein are significantly reduced, enhancing the resistance to change and the ability of DARTS protein decomposition.
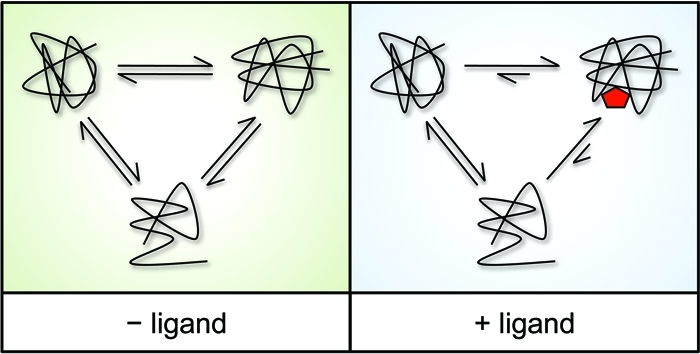
Figure 1. DARTS Principle [1]
The main experimental procedure of DARTS is to co-incubate small molecule drugs with proteins for a certain time, followed by adding protease for digestion. The binding of SM to target protein can make the protein more resistant to protease, after gel electrophoresis staining, compared to the non-drug digestion group, the protected bands are identified, then identified through mass spectrometry (MS).
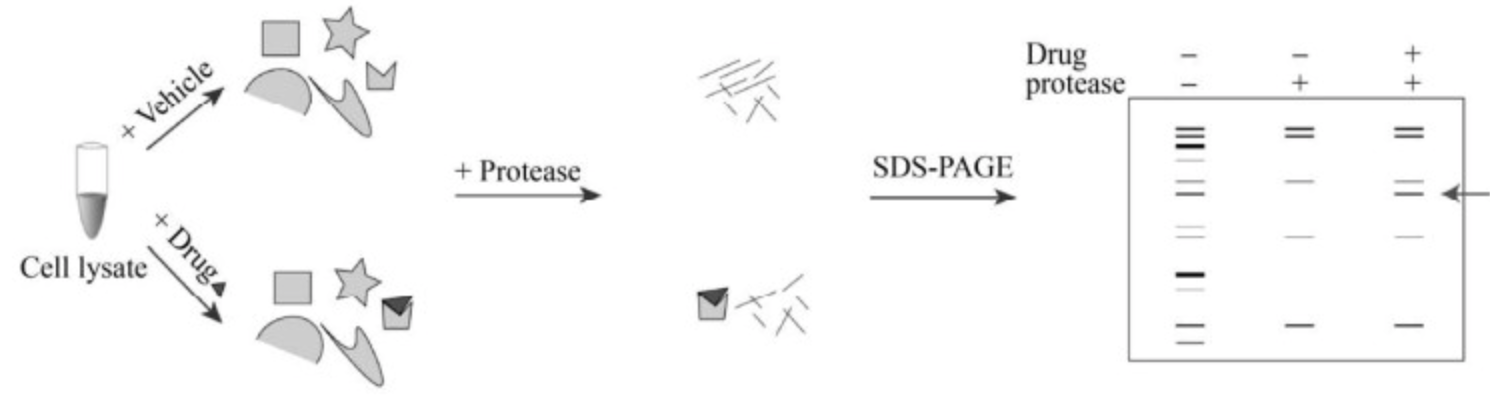
Figure 2. DARTS Experimental Process [2]
1. Protein Sample Preparation
Drug target discovery is a non-targeted process based on the study of all proteins in tissues or cells. DARTS can be used to screen various proteins, including different cell lines, blood, and tissues. Cell lines have better repeatability and adjustable protein abundance and are usually used as primary protein sources. Preparing high-quality protein samples is the basis for ideal results. The protein extraction process should be as simple as possible to avoid the loss of target proteins.
Protein extraction requires gentle and effective non-denaturing conditions to maintain their original structure when binding with SMs. To preserve protein structure, violent lysis (such as ultrasonication, mechanical breaking) or strong detergents (such as SDS lysis buffer, RIPA lysis buffer) should be avoided. DARTS usually recommends using some mild detergents (such as M-PER or TritonX-100) to extract water-soluble proteins without damaging organelle membranes. It is noteworthy that temperature plays a crucial role in DARTS, and the entire protein extraction process must be carried out on ice to avoid disrupting the cellular environment or causing unplanned modifications, structural folding, or degradation, which may lead to false positives. Additionally, appropriate protease or phosphatase inhibitors can be added to the lysis buffer to prevent protein degradation.
2. Binding of SMs and Proteins
To simulate the binding of SMs and proteins in vivo, SMs are added to the protein samples for incubation. The cell lysate is evenly divided into control and small molecule drug treatment groups, with protein concentrations typically in the range of 2.5-5 μg/μL. Then, the stock solution of SMs (usually 100 mM) is diluted in a gradient to the reaction solution, followed by incubation with proteins at room temperature for 30-60 minutes with moderate shaking. The incubation time depends on SMs and can be optimized accordingly. Although most SM-protein binding can be completed in seconds, they are usually incubated for at least 30 minutes to ensure sufficient binding.
The reaction concentration of SMs varies and usually depends on preliminary experiments. The basic trial can refer to the half-maximal inhibitory concentration (IC50) or half-maximal effective concentration (EC50) in biological assays. When conducting unbiased DARTS, it is recommended to use a small molecule concentration about 5-10 times more than IC50 or EC50 value to ensure detectable binding between SMs and the target protein. However, if the IC50 or EC50 has not been determined before DARTS, in most reported cases, a small molecule concentration of 100 μM is usually used. In addition, some experiments directly set the small molecule concentration to 250 μM to expand the binding results, which may increase the possibility of false positives yet.
The incubation temperature of SM-protein compound affects its binding effect and is usually set according to the stability of the protein. Although 37°C is the optimal temperature for simulating SM-protein binding, some temperature-sensitive proteins, such as α-synuclein, are easily degraded if they lose balance, resulting in a significant decrease in protein abundance. Moreover, setting at 0-4°C helps prevent premature degradation of proteins but slows down the process of SM-protein binding, which usually requires a longer cultivation time (4-6 hours). Therefore, room temperature (about 25°C) is a compromise choice for binding experiments and has been usually chosen in experiments. In addition, the solvent for the small molecule solution is preferably water, but if the SM is soluble in dimethyl sulfoxide (DMSO), it is necessary to ensure that the final concentration of DMSO has little effect on the biological state of the protein.
3. Protein Hydrolysis
After the binding experiment is completed, protease hydrolysis is required to distinguish the target protein from non-target proteins based on stability differences. Proteases need to be diluted in 1×TNC buffer, preferably prepared immediately for fresh use to avoid reduced protease activity. The total volume of 100 μL of the SM-protein complex is evenly transferred into several new tubes, followed by adding 2 μL of diluted protease solution for hydrolysis. Then, 2 μL of protease inhibitor is added to each tube for a incubation on ice for 10 minutes to stop digestion.
Optimized protease hydrolysis can fully digest proteins, which can eliminate background interference of non-target protein and provide identifiable SM-binding fragments for subsequent separation. Protein hydrolysis is a key step in the DARTS. For example, excessive hydrolysis usually breaks down the SM-target protein compound into small peptides, while insufficient hydrolysis will show no obvious difference in proteins. The appropriate concentration of protease is crucial for the DARTS. In most conditions, a feasible ratio of protease to protein ranges from 1:100 to 1:1000. Since various proteins expressed in cells have different sensitivities to proteases, it is recommended to optimize the ratio of protease to protein preliminarily. First, based on the protein concentration detected by the BCA assay, prepare preset solutions of different protease-to-protein ratios (for example, from 1:100 to 1:1000) to ensure that peptides/proteins can be well observed within a broad range. The concentration of protease can be further adjusted to visualize the interaction of SM-binding proteins.
4. SDS-PAGE Based Separation
In the DARTS experiment, protease hydrolysates are generally separated by their molecular size through SDS-PAGE. First, the hydrolysates are denatured at high temperatures. Then 10-20μL protein sample is added to the lanes. And gel electrophoresis is performed at constant voltage at room temperature until the dye front reaches the bottom of the gel. Finally, SDS-PAGE can be visualized by staining techniques.
Coomassie blue staining and silver staining are two commonly used staining methods. Coomassie blue staining can stain most proteins (5-25 ng) with high visibility, simple steps, including gel washing, staining, and destaining; but this method is time-consuming and difficult to detect low-concentration proteins. Coomassie blue dye will temporarily chemically modify the target protein and interact through non-covalent forces; therefore, stained gel bands can be completely faded, and the recovered peptide segments are easily collected for mass spectrometry (MS). In contrast, silver staining (including gel fixing, Na2S2O3 sensitization, AgNO3 coloring, and Na2CO3 development) is quick and sensitive, capable of detecting low-concentration proteins (0.25-0.5 ng). It is worth mentioning that common silver staining often use glutaraldehyde or formaldehyde as enhancers, which may cause chemical cross-linking of proteins in the gel matrix, affecting peptide analysis in MS. Therefore, the compatibility of silver staining kits with MS analysis should be considered.
5. Collection of Target Protein Gel Bands
After separating and visualizing the proteins on the gel image, SM-binding fragments can be captured by comparing the band density of the drug group and the control group. Due to increased stability, the bound proteins usually show thicker bands. Gel fragments are placed on a clean flat surface, and the stained areas of the target bands are cut out from the gel for LC-MS/MS. Usually, different bands can be visualized under illumination. If possible, it is best to use gel image processing tools, such as charge-coupled device (CCD) cameras and fluorescence scanners, for gel analysis to accurately identify differential bands.
6. Identification and Verification of Target Proteins
The rapid, sensitive, and available mass spectrometry databases support the discovery and identification of proteins/ peptides in drug research. They are usually used in unbiased DARTS methods for target identification. For MS analysis, after a series of in-gel enzymatic procedures, the extracted peptides are loaded onto a C18 reverse phase column for separation, and high-resolution MS data is collected. The target protein is identified in search of these protein database. Once potential targets are successfully identified by MS, it is recommended to verify the protein through Western blotting (WB), co-immunoprecipitation (Co-IP), or other experiments. WB are usually used to detect the accuracy of protein identification through antibody-antigen binding and SDS-PAGE separation, compared to membrane blot proteins to estimate the content of potential targets. Co-IP is a classical method for directly preparing proteins from cell lysate complexes, based on the specific interaction between antibodies and antigens. Thus, the correct proteins identified by LC-MS can be captured by their antigens, helping to eliminate incorrect identifications.
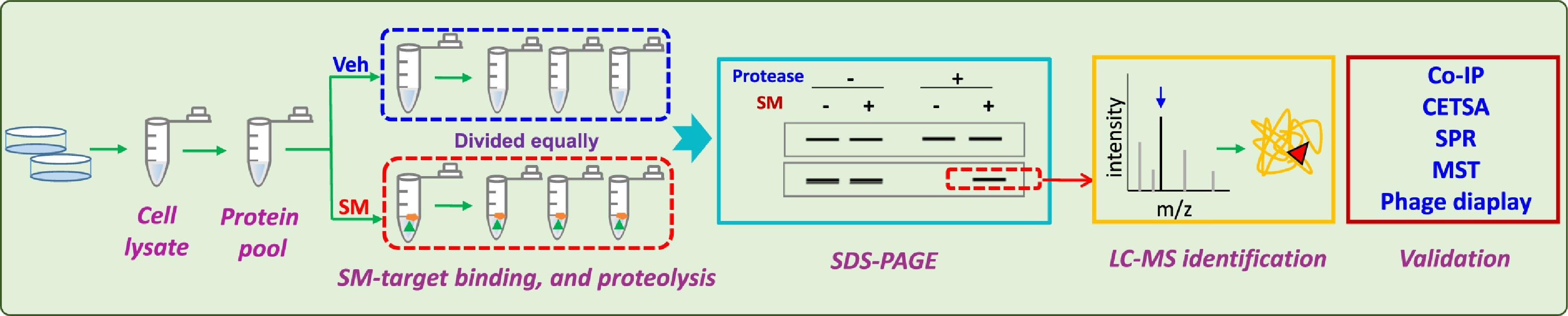
Figure 3. DARTS Experimental Process [2]
Analysis Workflow
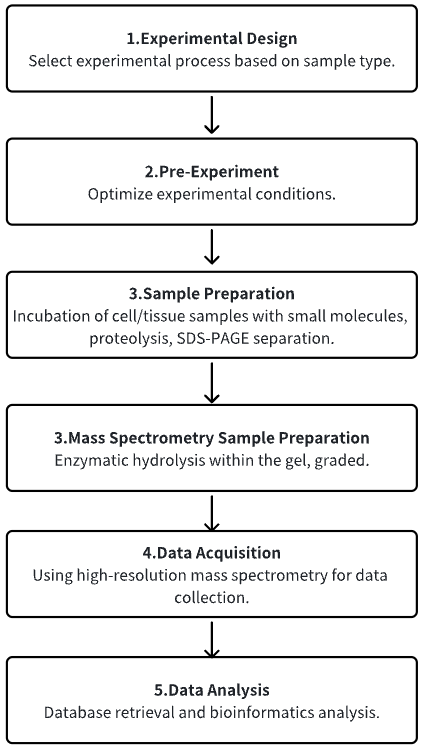
1. Determine the Experimental Process According to the Experimental Requirements
2. Pre-Experiment, Optimizing Experimental Conditions
3. Cell/Tissue Smples Are Incubated with Small Molecules and Separated
4. MS Sample Preparation
5. High-Resolution MS Acquisition Data
6. Data Retrieval and Analysis
Service Advantages
1. Identification of Target Proteins from Multiple Types of Sample Sources, etc.
2. Provide Guidance on Optimizing Experimental Conditions
3. High-Confidence, High-Precision MS Detection
4. Comprehensive Bioinformatics Analysis
Sample Results
1. Hydroxy-α-Sanshool from Pepper Fruit Induces Browning of White Adipose Tissue (WAT) by Activating TRPV1 and Causing PPAR-γ Deacetylation
Accumulating evidence suggest that increasing energy expenditure is a viable strategy to combat obesity, and promoting the browning of white adipose tissue (WAT) might be one attractive method. Hydroxy-α-sanshool (HAS), a natural amide alkaloid extracted from pepper fruit, has many benefits in lipid metabolism regulation. Studies have been conducted using obese animal models and 3T3-L1 differentiated cell models to investigate the anti-obesity effects of HAS. Various methods including micro-CT, metabolic cage assays, cellular mitochondrial stress tests, transmission electron microscopy, and cold exposure experiments were used to study the effects of HAS on the overall fat and liver in obese mice, and its role in inducing WAT browning. Methods such as real-time fluorescence quantitative PCR (qPCR), digital PCR (dPCR), immunoblotting, Co-IP, molecular docking, DARTS, and thermal shift assay were used to study the targets and mechanisms of HAS. The research found that treatment with HAS helped mice combat high-fat diet (HFD)-induced obesity and improved metabolic features. Moreover, The results indicated that the anti-obesity effects of HAS were related to increased energy expenditure and heat production by inducing WAT browning. Further studies revealed that HAS can upregulate the expression of UCP-1, increase the number of mitochondria, and enhance the cellular oxygen consumption rate (OCR) of white adipocytes. Importantly, the results showed that the browning effect of HAS was closely related to the activation of the TRPV1/AMPK pathway and SIRT1-dependent PPAR-γ deacetylation, with TRPV1 being a potential pharmaceutical target for the browning effects of HAS on WAT.
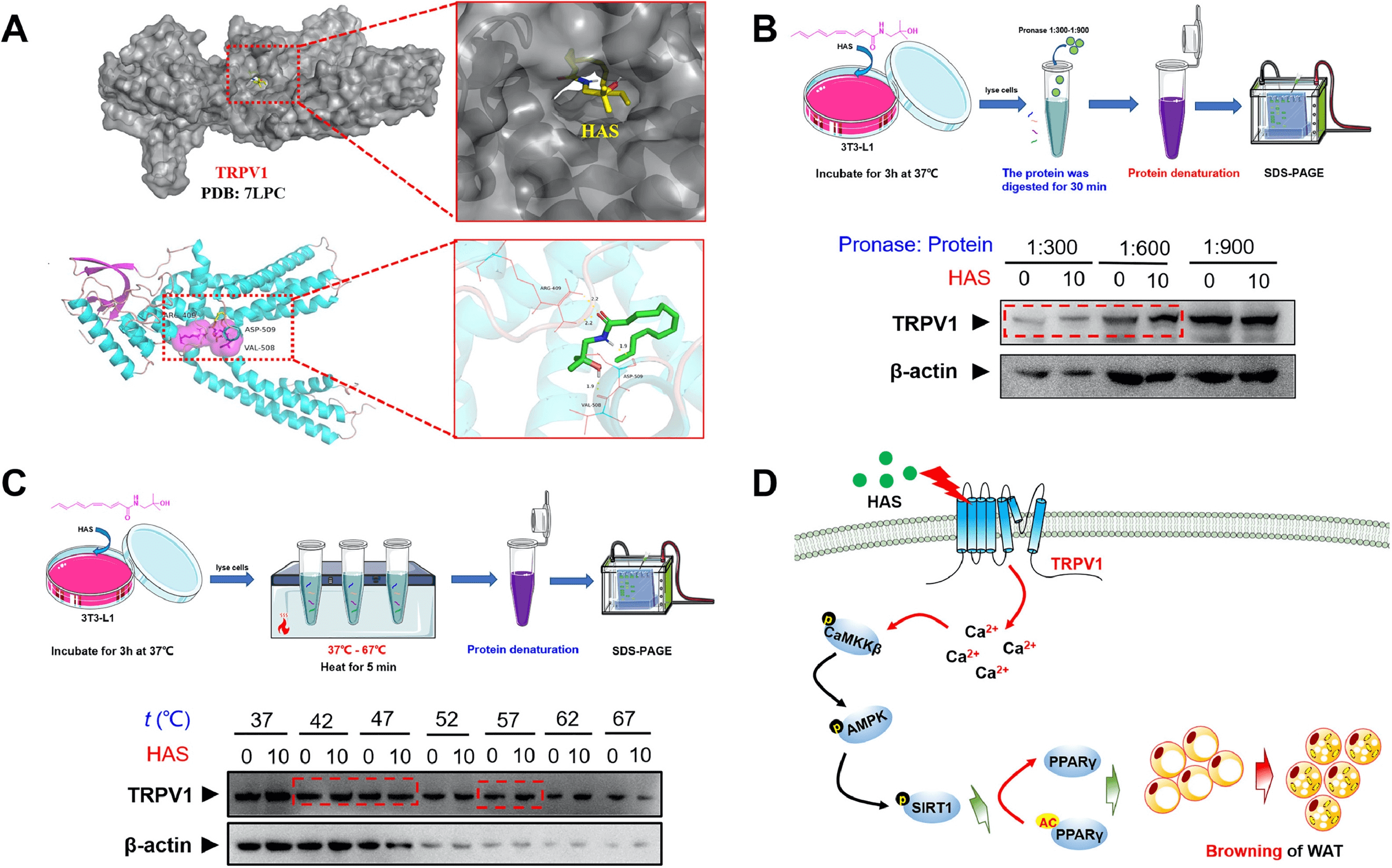
Figure 4. Study Drug Targets Through Molecular Docking, DARTS, and CESTA [3]
2. Luteolin Improves DSS-Induced Colitis in Mice by Inhibiting Macrophage Activation and Chemotaxis
Luteolin is known for its multifaceted therapeutic properties against inflammatory diseases and has the potential to address the unmet need for effective treatment of ulcerative colitis (UC), a common subtype of inflammatory bowel disease (IBD). A research aimed to comprehensively evaluate the therapeutic effects of luteolin in a dextran sulfate sodium (DSS)-induced colitis mouse model, revealing its anti-UC mechanisms. The study included rigorous histopathological examinations and biochemical analyses to assess the in vivo therapeutic potential of luteolin against DSS-induced colitis. The anti-inflammatory capabilities of luteolin were carefully examined in vitro using lipopolysaccharide (LPS)-stimulated RAW264.7 cells and primary peritoneal macrophages. Transwell and Zigmond chamber assays were used to quantitatively assess the effects of luteolin on C-C motif chemokine ligand 2 (CCL2)-induced macrophage migration. Additionally, methods such as thermal shift assay, DARTS, and molecular docking were used to identify the potential therapeutic targets of luteolin and study its binding sites and interaction modes. The results showed that luteolin improved symptoms of colitis, restored intestinal barrier integrity, and inhibited the production of pro-inflammatory cytokines in colonic tissue, demonstrating its potential to combat DSS-induced colitis. Moreover, luteolin displayed strong anti-inflammatory activity in vitro in LPS-stimulated RAW264.7 cells and primary peritoneal macrophages. Notably, luteolin inhibited the phosphorylation of IKKα/β, IκBα, and p65, while preventing the degradation of IκBα in LPS-treated RAW264.7 cells and peritoneal macrophages. Furthermore, luteolin impaired the migration behavior of RAW264.7 cells and peritoneal macrophages, as demonstrated by the reduced migration distance and speed of luteolin-treated macrophages. Mechanistically, luteolin antagonized IKKα/β, subsequently inhibiting the phosphorylation of IKKα/β and the activation of NF-κB signaling.
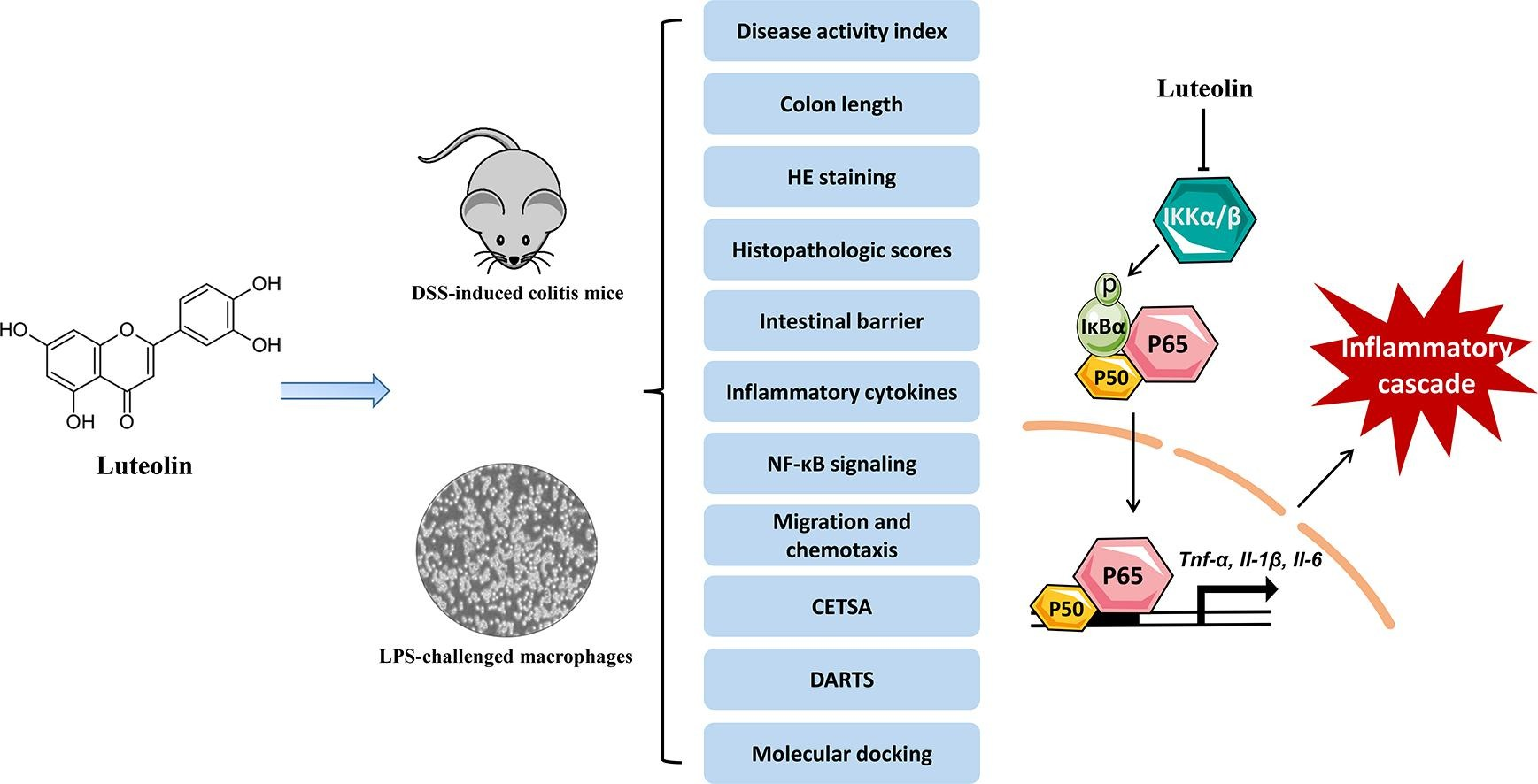
Figure 5. Research on the Target Mechanism of Luteolin [4]
3. Narirutin Improves Alcohol-Induced Liver Injury in Zebrafish Larvae by Targeting MAPK14
Alcohol-related liver disease (ALD) encompasses a range of hepatic abnormalities, including isolated alcoholic steatosis, steatohepatitis, and cirrhosis. The flavanone-7-O-glycoside narirutin (NRT), a major flavonoid in citrus peel, exhibits antioxidant, anti-inflammatory, and lipid-lowering activities. Research has explored the effects of NRT on alcohol-induced liver injury by alcohol and its potential mechanisms. Using zebrafish larvae, NRT's impact on acute exposure to ethanol (EtOH) was studied. Liver phenotypic, morphological, and biochemical assessments were conducted to evaluate NRT's hepatoprotective effects. Through network pharmacology and molecular docking analysis, potential targets of NRT in EtOH-induced liver injury were identified. DARTS was used to evaluate the binding of NRT to mitogen-activated protein kinase 14 (MAPK14). Reverse transcription quantitative real-time PCR (RT-qPCR) and Western blotting (WB) were conducted to verify NRT's mechanisms of action. Liver phenotypic, morphological, and biochemical assessments showed that NRT had potential therapeutic effects against acute EtOH-induced liver injury. RT-qPCR confirmed that NRT reversed changes in gene expressions related to oxidative stress, lipid production, and the endoplasmic reticulum (ER)/unfolded protein response pathways. Network pharmacology and molecular docking analysis identified NRT's potential targets and confirmed that NRT regulates the p38 MAPK signaling pathway by targeting MAPK14.
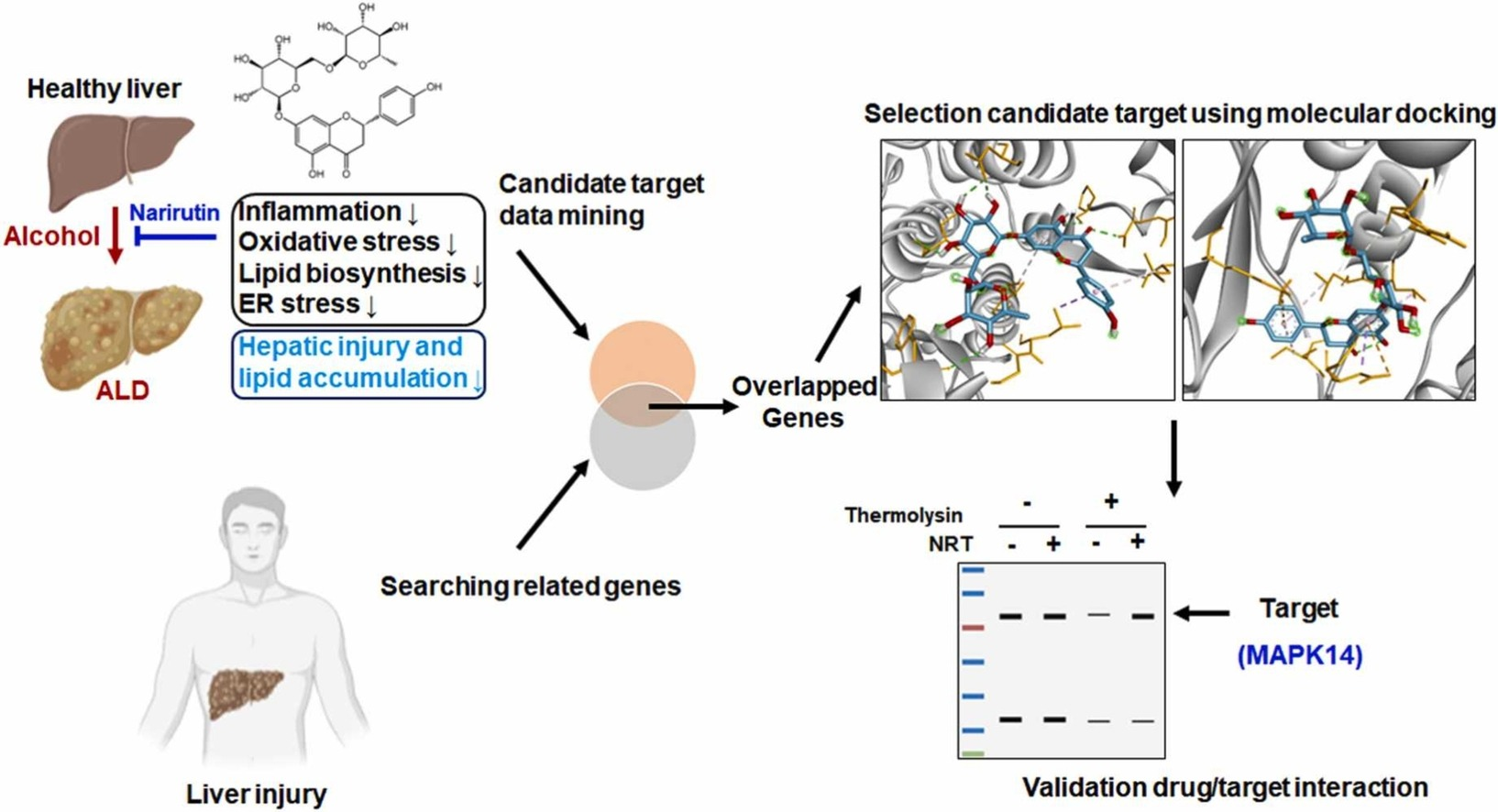
Figure 6. Evaluation of the Binding of NRT to MAPK14 Using DARTS [5]
Sample Submission Requirements
1. Samples such as drugs/proteins should not be contaminated with impurities as much as possible.
Services at MtoZ Biolabs
1. The Complete Experimental Procedure
2. Relevant Instrument Parameters
3. Raw MS Data
4. Data Analysis Reports
Applications
1. Discovery of Drug Targets
In recent years, many researchers have adopted DARTS as a method for screening drug targets or validating drug-protein binding, with DARTS being favored in the fields of anticancer and antitumor drug research. Additionally, DARTS has been extensively applied to elucidate the pharmacological mechanisms of action in anti-neurodegenerative, anti-angiogenic, anti-inflammatory, antifungal and antitoxin diseases.
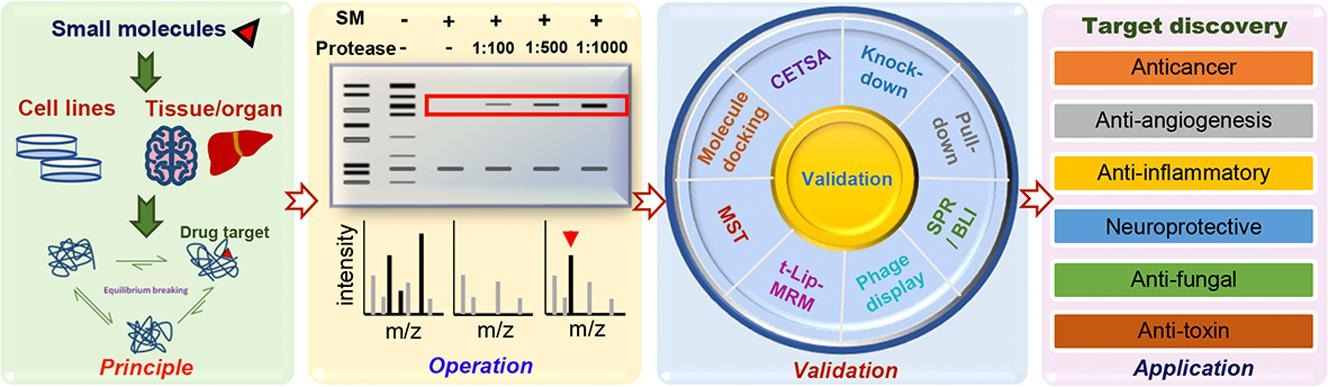
Figure 7. Applications of DARTS [2]
2. Development of New Methods
In silico molecular docking between SMs and target proteins is a predictive method to determine the preferred orientation of stable complexes. Molecular docking is one of the most practical methods to predict a reasonable binding conformation between bioactive small molecule ligand and binding target proteins. The binding of bioactive small molecule ligand to the protein sites can be described through several high-affinity interactions to determine its most stable structure. These interactions include hydrophobic interactions, electrostatic interactions, and hydrogen bonding interactions. A comprehensive analysis can be performed on stable peptide sequences determined by DARTS and LC-MS/MS, as well as amino acids that directly interact with bioactive SMs determined by in silico molecular docking. These amino acids can provide key binding groups, offering essential information for biological and phenotypic validation mutation studies.
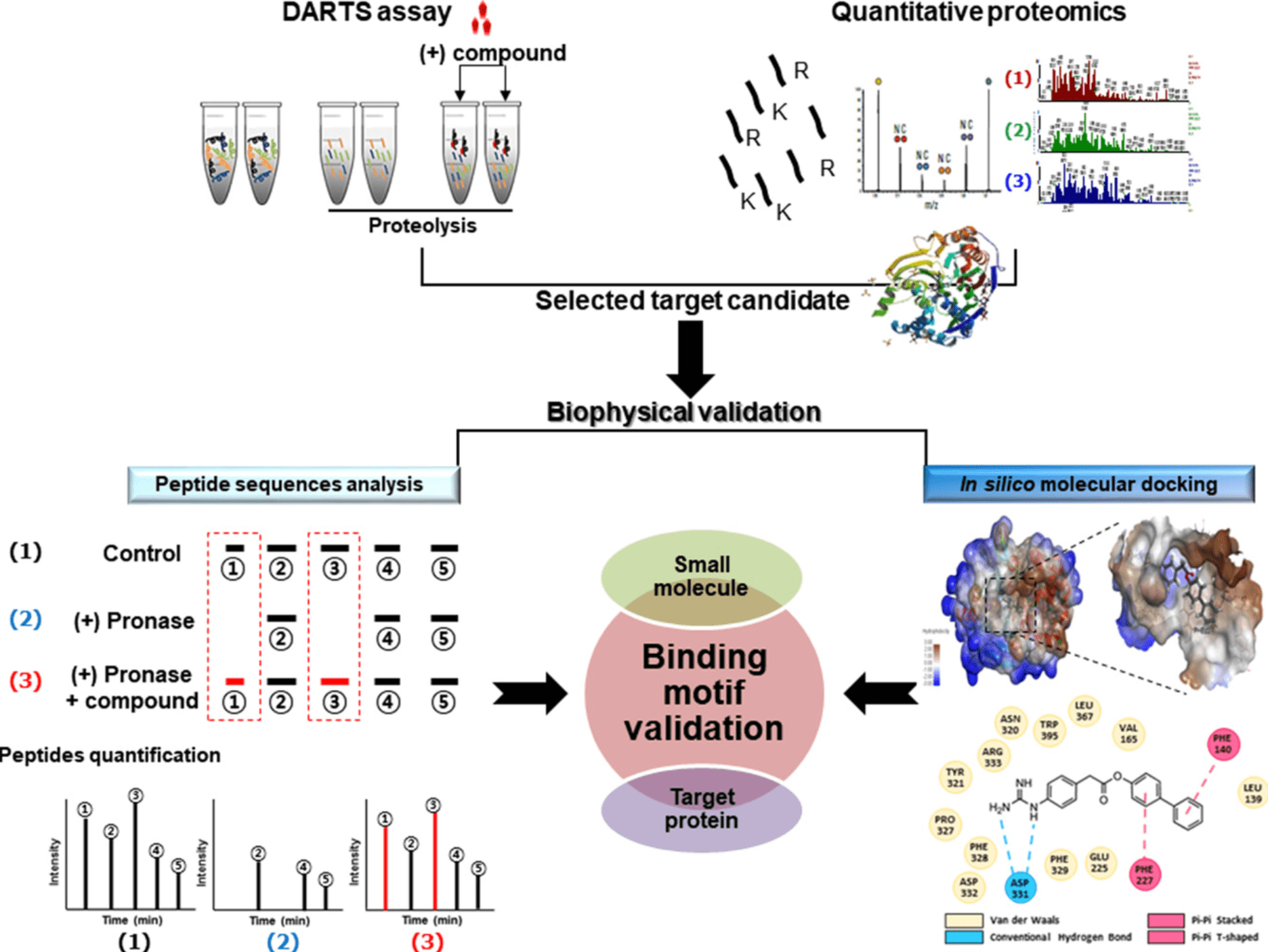
Figure 8. Molecular Docking Combined with DARTS [6]
FAQ
Q1: What are the advantages and disadvantages of DARTS?
DARTS has several advantages: (1) It does not require chemical modification of protein. (2) It can discover new targets in single-component SMs as well as natural complex components. (3) It does not require protein purity. (4) It can determine the main binding groups between bioactive SMs and their target proteins. However, DARTS also has certain limitations: (1) It can be performed in vitro, but is challenging in vivo. (2) Traditional DARTS method relies on SDS-PAGE and gel staining for visual observation, while the abundance of most proteins in cells and tissues is low, which makes it impossible to detect the binding target proteins of SMs through unbiased proteomics. (3) Some proteins are insensitive to proteases under native conditions, which confounds isolation of true targets.
References
[1] Lomenick B, Olsen RW, Huang J. Identification of direct protein targets of small molecules. ACS Chem Biol. 2011 Jan 21;6(1):34-46. doi: 10.1021/cb100294v. Epub 2010 Nov 30. PMID: 21077692; PMCID: PMC3031183.
[2] Ren YS, Li HL, Piao XH, Yang ZY, Wang SM, Ge YW. Drug affinity responsive target stability (DARTS) accelerated small molecules target discovery: Principles and application. Biochem Pharmacol. 2021 Dec;194:114798. doi: 10.1016/j.bcp.2021.114798. Epub 2021 Oct 19. PMID: 34678227.
[3] Zhang Q, He CX, Wang LY, Qian D, Tang DD, Jiang SN, Chen WW, Wu CJ, Peng W. Hydroxy-α-sanshool from the fruits of Zanthoxylum bungeanum Maxim. promotes browning of white fat by activating TRPV1 to induce PPAR-γ deacetylation. Phytomedicine. 2023 Dec;121:155113. doi: 10.1016/j.phymed.2023.155113. Epub 2023 Sep 21. PMID: 37748388.
[4] Xue L, Jin X, Ji T, Li R, Zhuge X, Xu F, Quan Z, Tong H, Yu W. Luteolin ameliorates DSS-induced colitis in mice via suppressing macrophage activation and chemotaxis. Int Immunopharmacol. 2023 Nov;124(Pt B):110996. doi: 10.1016/j.intimp.2023.110996. Epub 2023 Sep 29. PMID: 37776768.
[5] Park KH, Makki HMM, Kim SH, Chung HJ, Jung J. Narirutin ameliorates alcohol-induced liver injury by targeting MAPK14 in zebrafish larvae. Biomed Pharmacother. 2023 Oct;166:115350. doi: 10.1016/j.biopha.2023.115350. Epub 2023 Aug 24. PMID: 37633055.
[6] Hwang HY, Kim TY, Szász MA, Dome B, Malm J, Marko-Varga G, Kwon HJ. Profiling the Protein Targets of Unmodified Bio-Active Molecules with Drug Affinity Responsive Target Stability and Liquid Chromatography/Tandem Mass Spectrometry. Proteomics. 2020 May;20(9):e1900325. doi: 10.1002/pmic.201900325. Epub 2020 Feb 5. PMID: 31926115.
How to order?







