Untargeted Metabolomics Service
Untargeted metabolomics is a study in which all detectable metabolites are identified and quantitated for analyzing the overall metabolic characteristics and discovering metabolic mechanisms behind the phenomenon. Also known as discovery metabolomics, untargeted metabolomics is a powerful tool for large-scale early phase diagnosis and discovery of new biomarkers. Compared to traditional metabolic quantitation methods, untargeted metabolomics is a high-throughput technology, enabling more comprehensive analysis with more accurate data acquisition, eliminating biased research direction.
Untargeted Metabolomics

MtoZ Biolabs is proud to offer high-efficient and accurate plant metabolomics analysis service, based on Thermo Q Exactive and AB Q-TOF 5600 mass spectrometry systems.

MtoZ Biolabs develops suitable microbial sample preparation and metabolites extraction strategies according to your projects, and assures high-efficient and accurate microbial metabolomics analysis.
Analysis Workflow
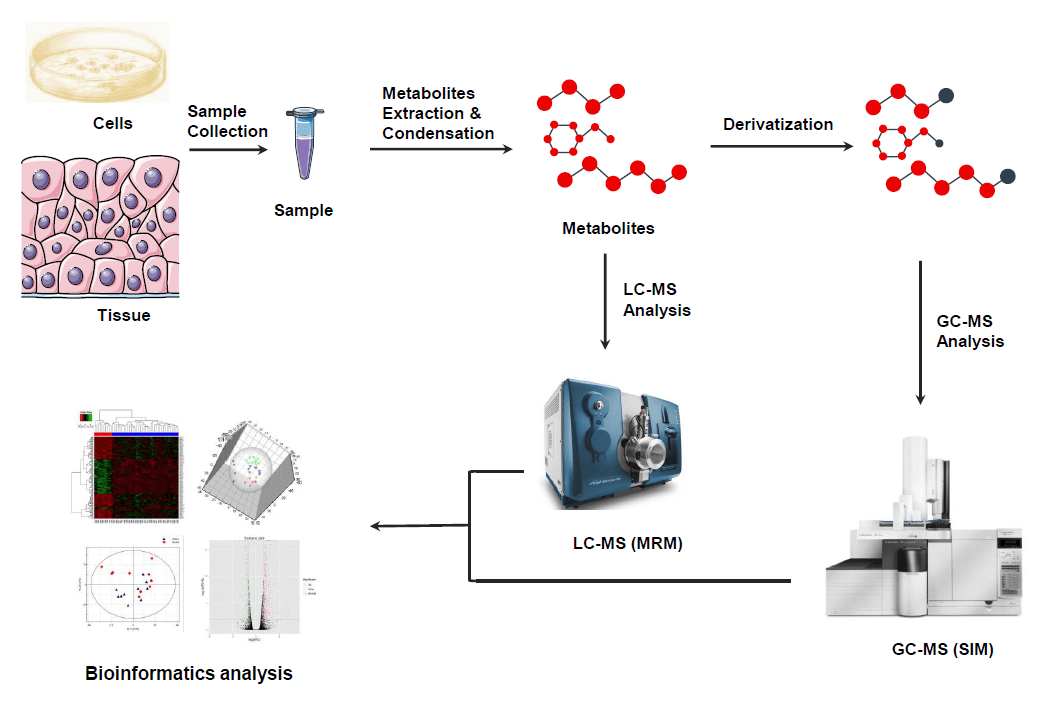
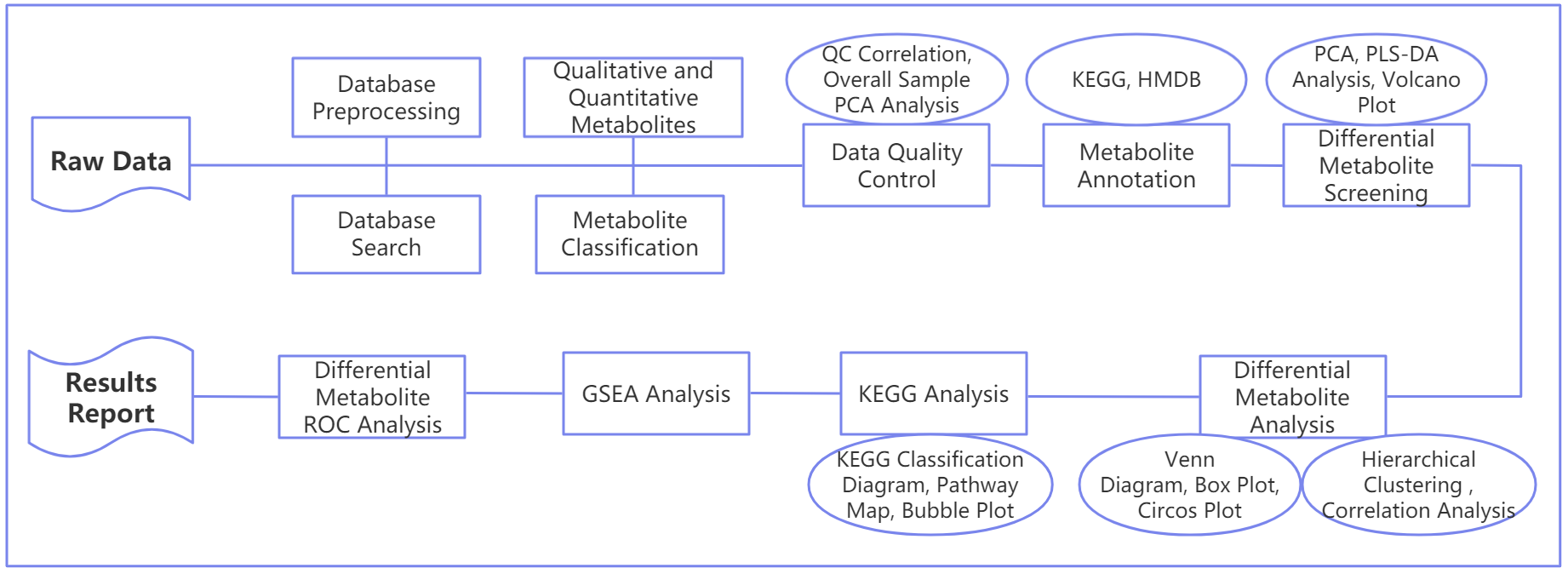
Data Analysis
Service Advantages
1. Standard Technical Platform
2. State-of-the-Art LC-MS and GC-MS Analysis Systems
3. Comprehensive Commercial Databases System
4. Professional Support and Precise Custom Service
Sample Types
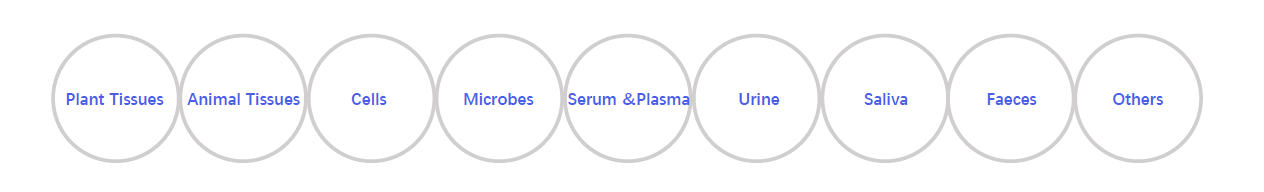
Sample Submission Requirements
1. Cells and Microbes Samples
1×10^7 cells/sample. Cellular activities should be terminated instantly, whereas the cells remain intact.
2. Animal Tissues
100 mg/sample. Collect soft tissues like brain, heart, liver, muscle, and skin. Immediately add preservative reagent and freeze at -80°C.
3. Plant Tissues
200 mg/sample. Samples should be frozen in liquid nitrogen right after sample collection, and then transferred to -80 °C for storage.
4. Serum Samples
Repeated freezing and thawing of sample must be eliminated. Serum samples should be settled down at room temperature for 30 min in the collection tube, and then transferred to centrifuge tube and centrifuged at 8,000 rpm for 5 min. After centrifugation, supernatant is aliquoted to freezing tubes with 200 uL/sample, and stored at -80 °C.
5. Urine Samples
2 mL/sample. Urine samples can be aliquoted to centrifuge tubes with 2 mL each tube and addition of 1/100 (w/v) sodium azide, and stored at -80 °C.
6. Faeces Samples
200 mg/sample
Deliverables
1. Experiment Procedures
2. Parameters of Liquid Chromatography and Mass Spectrometer
3. MS Raw Data Files
4. MS Data Quality Checks
5. Metabolites Quantification Data
6. Bioinformatics Analysis (PCA, KEGG, etc)
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Exosome Untargeted Metabolomics Analysis Service
Animal Untargeted Metabolomics Service
Targeted Metabolomics Analysis Service
Metabolomics
Quantitative Proteomics
Protein Analysis
How to order?







