Protein Identification Service
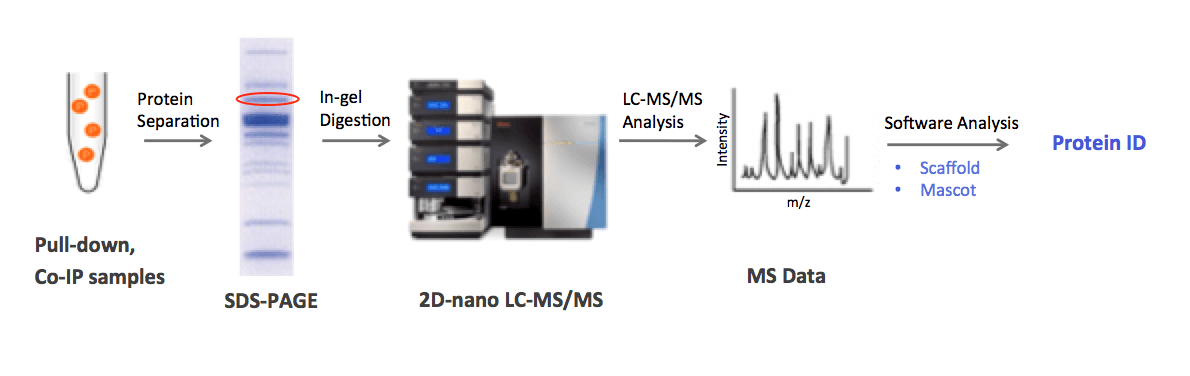
MtoZ Biolabs utilizes 2D-nano LC-MS/MS technology for protein identification. In our general workflow, protein complexes are separated in 1D/2D gel and the target protein is then digested into peptide fragments, followed by HPLC separation and tandem MS analysis. We use Scaffold and Mascot software for analyzing peptide MS data, and ensuring confident protein identification.
Experimental Instruments
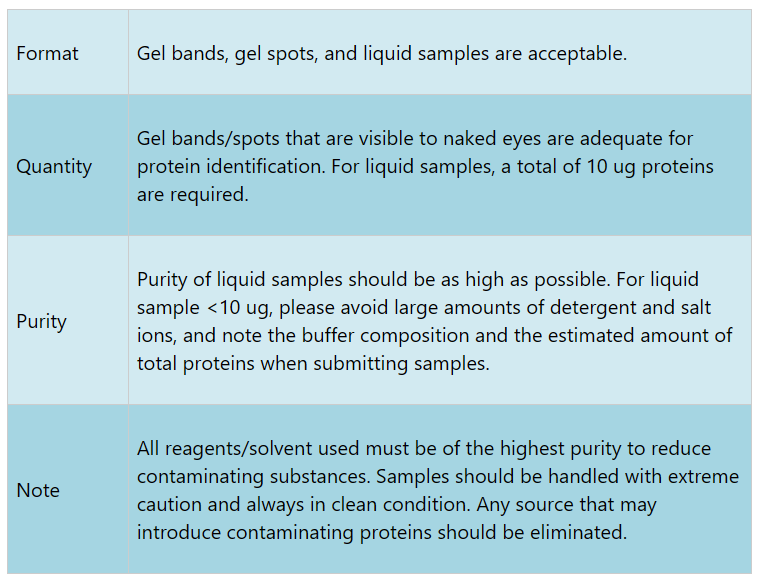
Sample Submission Requirements
Deliverables
1. Experiment Procedures
2. Parameters of Liquid Chromatography and Mass Spectrometer
3. MS Raw Data Files
4. Peptide Identifications and Intensity
5. Protein Identifications and Intensity
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Mass Spectrometry-Based Protein Identification Service
Protein Structure Identification Service
Unknown Protein Identification Service
Protein Analysis
LC-MS Analysis of Pull-Down Proteins
Protein Sequencing
Protein Full-Length Sequencing
PTM Analysis
Submit Inquiry
How to order?







