Multi Reaction Monitoring MRM Service
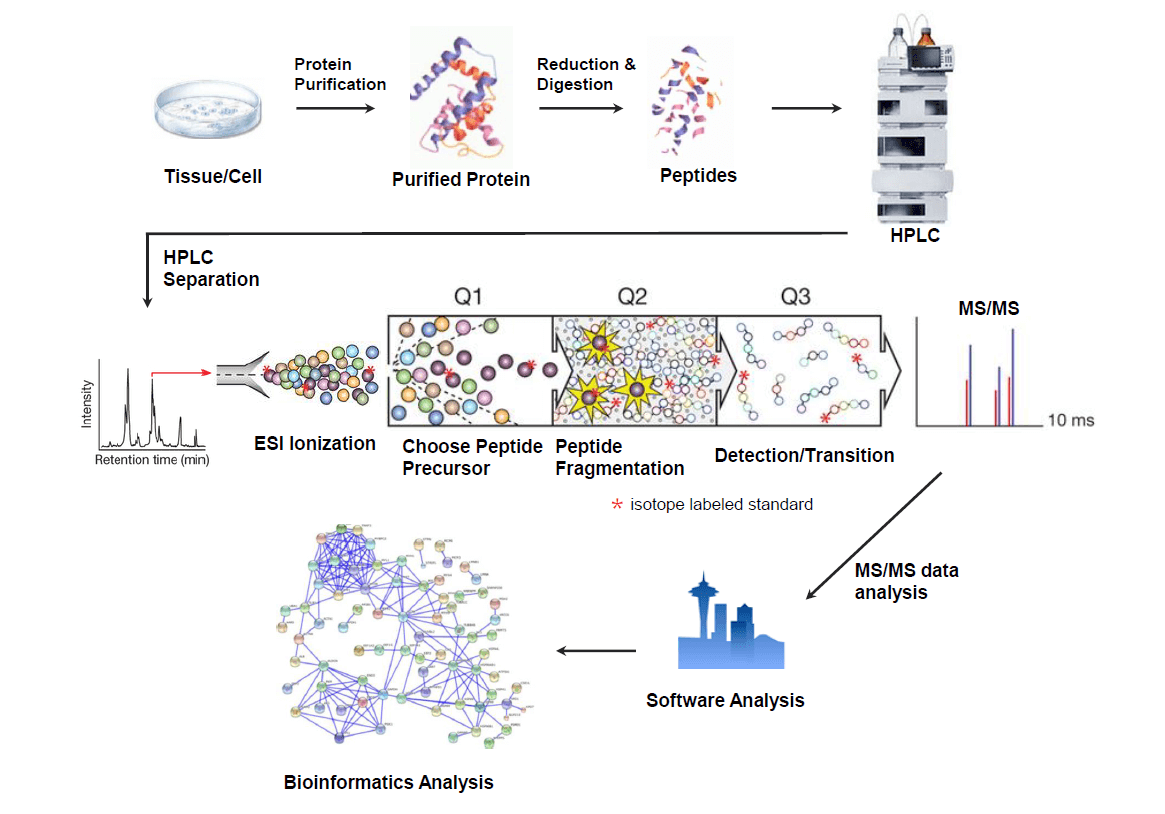
Multiple Reaction Monitoring (MRM) is a quantitative method for analyzing targeted proteomics. Based on the sequence information of targeted proteins, MRM selectively analyzes proteins correlating with the targeted peptide signals, eliminating signals from interfering peptides. There are three steps in MRM mass spectrometry analysis: isolation of targeted precursor peptides based on MS spectra, fragmentation of precursor peptides, and detection of fragmented ions based on MS/MS spectra.
MRM has the highest accuracy among all proteomics quantitation methods, enabling absolute quantitation of protein with reference to internal standard, and analysis of up to 200 proteins at the same time. MtoZ Biolabs is proud to offer an all-inclusive MRM service. Just tell us your project objective and send us the protein sample, we will perform all the corresponding experiments, and provide the most professional analytical service to meet your specific needs.
Analytical Platform
AB SCIEX TripleTOF 5600, AB SCIEX Triple Quad™ 5500 and Q Exactive Fusion
Analysis Workflow
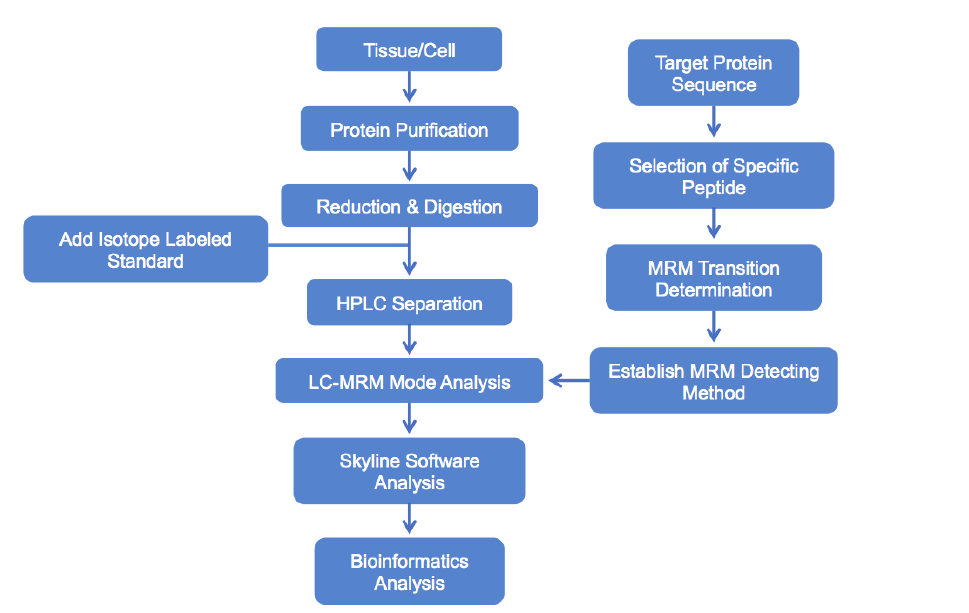
Service Advantages
1. Offers the Highest Specificity, Sensitivity, and Reproducibility for Proteomics Quantitation
2. Broad Dynamic Range, Covering 4 Orders of Magnitude of Protein Abundance
3. High-Throughput, and Can Identify Up to 200 Specific Proteins at Once
4. Absolute Quantitation of Multiple Proteins Without the Need of Antibody
Applications
1. Verification of Untargeted Proteomics Results, Such as Label-Free Proteomics
2. Absolute Quantitation of Up to 500 Proteins/Peptides
3. Analysis of Highly Homologous Protein Family Members
4. Protein Post-Translational Modification Analysis
5. Discovery and Absolute Quantitation of Biomarkers
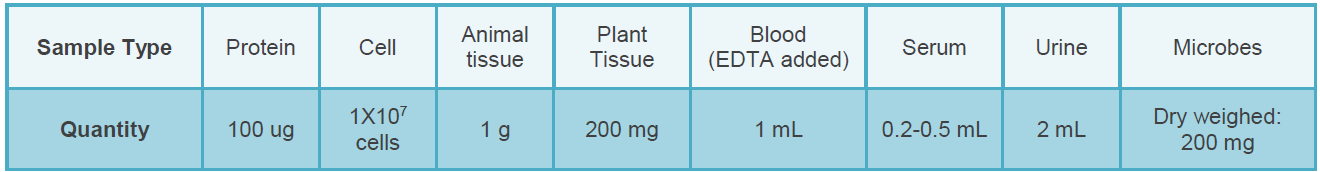
Sample Submission Requirements
Bioinformatics Analysis
1. MRM Data Quality Assessment
2. Multivariate PCA Analysis
3. Protein Statistical Analysis: Venn Diagram, Volcano Plot
4. Functional Annotation: GO annotation, KEGG Annotation, COG Annotation
5. Clustering Analysis: Hierarchical Clustering, K-means Clustering
6. Network Analysis: STRING Analysis
Deliverables
1. Experiment Procedures
2. Parameters of Liquid Chromatography and Mass Spectrometer
3. MS Raw Data Files
4. Peptide Identifications and Intensities
5. Protein Identifications and Intensities
6. Bioinformatics Analysis
Related Services
Label-Free
iTRAQ/TMT
SILAC/Dimethyl
SWATH
MRM
Protein Identification
Protein Mass Measurement
PTMs Identification
Phospho Proteomics
Acetyl Proteomics
Ubiquitin Proteomics
Glyco Proteomics
Disulfide Bond
Histone Modifications
How to order?







